A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana
- PMID: 15695592
- PMCID: PMC548969
- DOI: 10.1073/pnas.0406377102
A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana
Erratum in
- Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5635
Abstract
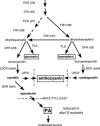
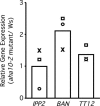
The plasma membrane in plant cells is energized with an electrical potential and proton gradient generated through the action of H+ pumps belonging to the P-type ATPase superfamily. The Arabidopsis genome encodes 11 plasma membrane H+ pumps. Auto-inhibited H+-ATPase isoform 10 (AHA10) is expressed primarily in developing seeds. Here we show that four independent gene disruptions of AHA10 result in seed coats with a transparent testa (tt) phenotype (light-colored seeds). A quantitative analysis of extractable flavonoids in aha10 seeds revealed an approximately 100-fold reduction of proanthocyanidin (PA), one of the two major end-product pigments in the flavonoid biosynthetic pathway. In wild-type seed coat endothelial cells, PA accumulates in a large central vacuole. In aha10 mutants, the formation of this vacuole is impaired, as indicated by the predominance of multiple small vacuoles observed by fluorescence microscopy using a vacuole-specific dye, 5-(and -6)-carboxy 2',7'-dichlorofluorescein diacetate. A similar vacuolar defect was also observed for another tt mutant, tt12, a proton-coupled multidrug and toxic compound extrusion transporter potentially involved in loading provacuoles with a flavonoid intermediate required for PA production. The endothelial cells in aha10 mutants are otherwise healthy, as indicated by the lack of a significant decrease in (i) the accumulation of other flavonoid pathway end products, such as anthocyanins, and (ii) mRNA levels for two endothelium-specific transcripts (TT12 and BAN). Thus, the specific effect of aha10 on vacuolar and PA biogenesis provides genetic evidence to support an unexpected endomembrane function for a member of the plasma membrane H+-ATPase family.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

