Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis
- PMID: 15696197
- PMCID: PMC546460
- DOI: 10.1172/JCI23523
Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis
Abstract
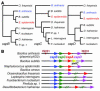
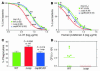
Coagulase-negative staphylococci, with the leading species Staphylococcus epidermidis, are the predominant cause of hospital-acquired infections. Treatment is especially difficult owing to biofilm formation and frequent antibiotic resistance. However, virulence mechanisms of these important opportunistic pathogens have remained poorly characterized. Here we demonstrate that S. epidermidis secretes poly-gamma-DL-glutamic acid (PGA) to facilitate growth and survival in the human host. Importantly, PGA efficiently sheltered S. epidermidis from key components of innate host defense, namely antimicrobial peptides and neutrophil phagocytosis, and was indispensable for persistence during device-related infection. Furthermore, PGA protected S. epidermidis from high salt concentration, a key feature of its natural environment, the human skin. Notably, PGA was synthesized by all tested strains of S. epidermidis and a series of closely related coagulase-negative staphylococci, most of which are opportunistic pathogens. Our study presents important novel biological functions for PGA and indicates that PGA represents an excellent target for therapeutic maneuvers aimed at treating disease caused by S. epidermidis and related staphylococci.
Figures


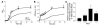
References
-
- Vuong C, Otto M. Staphylococcus epidermidis infections. Microbes Infect. 2002;4:481–489. - PubMed
-
- Götz F. Staphylococcus and biofilms. Mol. Microbiol. 2002;43:1367–1378. - PubMed
-
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. - PubMed
-
- Yao Y, Sturdevant DE, Otto M. Genome-wide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into S. epidermidis biofilm pathophysiology and the role of phenol-soluble modulins in biofilm formation. J. Infect. Dis. 2005;191:289–298. - PubMed
-
- Oppermann-Sanio FB, Steinbüchel A. Occurrence, functions and biosynthesis of polyamides in microorganisms and biotechnological production. Naturwissenschaften. 2002;89:11–22. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

