Genomic insights that advance the species definition for prokaryotes
- PMID: 15701695
- PMCID: PMC549018
- DOI: 10.1073/pnas.0409727102
Genomic insights that advance the species definition for prokaryotes
Abstract
To help advance the species definition for prokaryotes, we have compared the gene content of 70 closely related and fully sequenced bacterial genomes to identify whether species boundaries exist, and to determine the role of the organism's ecology on its shared gene content. We found the average nucleotide identity (ANI) of the shared genes between two strains to be a robust means to compare genetic relatedness among strains, and that ANI values of approximately 94% corresponded to the traditional 70% DNA-DNA reassociation standard of the current species definition. At the 94% ANI cutoff, current species includes only moderately homogeneous strains, e.g., most of the >4-Mb genomes share only 65-90% of their genes, apparently as a result of the strains having evolved in different ecological settings. Furthermore, diagnostic genetic signatures (boundaries) are evident between groups of strains of the same species, and the intergroup genetic similarity can be as high as 98-99% ANI, indicating that justifiable species might be found even among organisms that are nearly identical at the nucleotide level. Notably, a large fraction, e.g., up to 65%, of the differences in gene content within species is associated with bacteriophage and transposase elements, revealing an important role of these elements during bacterial speciation. Our findings are consistent with a definition for species that would include a more homogeneous set of strains than provided by the current definition and one that considers the ecology of the strains in addition to their evolutionary distance.
Figures

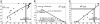



References
-
- Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., Moore, L. H., Moore, W. E. C., Murray, R. G. E., Stackebrandt, E., et al. (1987) Int. J. Syst. Bacteriol. 37, 463-464.
-
- Brenner, D., Staley, J. & Krieg, N. (2000) Bergey's Manual of Systematic Bacteriology (Springer, New York).
-
- Stackebrandt, E., Frederiksen, W., Garrity, G. M., Grimont, P. A. D., Kampfer, P., Maiden, M. C. J., Nesme, X., Rossello-Mora, R., Swings, J., Truper, H. G., et al. (2002) Int. J. Syst. Evol. Microbiol. 52, 1043-1047. - PubMed
-
- Rossello-Mora, R. & Amann, R. (2001) FEMS Microbiol. Rev. 25, 39-67. - PubMed
-
- Cohan, F. M. (2002) Annu. Rev. Microbiol. 56, 457-487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

