Perinuclear localization of slow troponin C m RNA in muscle cells is controlled by a cis-element located at its 3' untranslated region
- PMID: 15701732
- PMCID: PMC1370719
- DOI: 10.1261/rna.5460105
Perinuclear localization of slow troponin C m RNA in muscle cells is controlled by a cis-element located at its 3' untranslated region
Abstract
The process of mRNA localization within a specific cytoplasmic region is an integral aspect of the regulation of gene expression. Furthermore, colocalization of mRNAs and their respective translation products may facilitate the proper assembly of multi-subunit complexes like the thick and thin filaments of muscle. This postulate was tested by investigating the cytoplasmic localization of three mRNAs-the alpha-actin, slow troponin C (sTnC), and slow troponin I (sTnI), which encode different poly-peptide partners of the thin filament. Using in situ hybridization we showed that all three thin filament mRNAs are localized in the perinuclear cytoplasm of cultured C2C12 muscle cells. Their localization differs from that of the nonmuscle beta-actin mRNA, which is localized in the peripheral region of both proliferating nondifferentiated myoblasts and the differentiated myocytes. Analysis of the localization signal of the sTnC mRNA showed that a 40-nucleotide-long region of the sTnC mRNA 3' UTR is sufficient to confer the perinuclear localization on a heterologous reporter beta-Gal mRNA. This localization signal showed tissue specificity and worked only in the differentiated myocytes, but not in the proliferating myoblasts or in HeLa cells. The predicted secondary structure of the localization signal suggests the presence of multiple stem and loop structures in this region of the 3' UTR. Mutations within the stem region of the localization signal, which abolish the base pairing in this region, significantly reduced its perinuclear mRNA localization activity. Using UV-induced photo-cross-linking of RNA and proteins we found that a myotube-specific 42-kDa polypeptide binds to the localization signal.
Figures
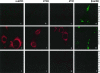

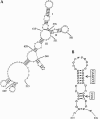
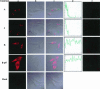
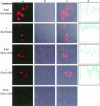
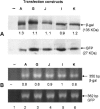
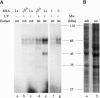
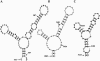
Similar articles
-
An AU-rich stem-loop structure is a critical feature of the perinuclear localization signal of c-myc mRNA.Biochem J. 2005 Dec 15;392(Pt 3):475-83. doi: 10.1042/BJ20050812. Biochem J. 2005. PMID: 16042622 Free PMC article.
-
Characterization of the cis-acting element directing perinuclear localization of the metallothionein-1 mRNA.Biochem Soc Trans. 2004 Nov;32(Pt 5):702-4. doi: 10.1042/BST0320702. Biochem Soc Trans. 2004. PMID: 15493992 Review.
-
Possible Cis-acting signal that could be involved in the localization of different mRNAs in neuronal axons.Theor Biol Med Model. 2005 Aug 24;2:33. doi: 10.1186/1742-4682-2-33. Theor Biol Med Model. 2005. PMID: 16120223 Free PMC article.
-
An eleven nucleotide section of the 3'-untranslated region is required for perinuclear localization of rat metallothionein-1 mRNA.Biochem J. 2005 Apr 15;387(Pt 2):419-28. doi: 10.1042/BJ20040630. Biochem J. 2005. PMID: 15537387 Free PMC article.
-
Determinants of mRNA localization.Curr Opin Cell Biol. 1992 Dec;4(6):975-8. doi: 10.1016/0955-0674(92)90128-y. Curr Opin Cell Biol. 1992. PMID: 1485968 Review.
Cited by
-
A systematic analysis of disease-associated variants in the 3' regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3' UTR variants.Hum Genet. 2006 Oct;120(3):301-33. doi: 10.1007/s00439-006-0218-x. Epub 2006 Jun 29. Hum Genet. 2006. PMID: 16807757 Review.
-
Hairpin structure within the 3'UTR of DNA polymerase beta mRNA acts as a post-transcriptional regulatory element and interacts with Hax-1.Nucleic Acids Res. 2007;35(16):5499-510. doi: 10.1093/nar/gkm502. Epub 2007 Aug 17. Nucleic Acids Res. 2007. PMID: 17704138 Free PMC article.
-
Nuclear tropomyosin and troponin in striated muscle: new roles in a new locale?J Muscle Res Cell Motil. 2013 Aug;34(3-4):275-84. doi: 10.1007/s10974-013-9356-7. Epub 2013 Aug 2. J Muscle Res Cell Motil. 2013. PMID: 23907338 Review.
References
-
- Ausubel, F.F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smitz, J.A., and Struhl, K. 1994. Current protocols in molecular biology. Greene and Wiley-Interscience, New York.
-
- Bally-Cuif, L., Schatz, W.J., and Ho, R.K. 1998. Characterization of the zebrafish Orb/CPEB-related RNA binding protein and localization of maternal components in the zebrafish oocyte. Mech. Dev. 77: 31–47. - PubMed
-
- Bashirullah, A., Cooperstock, R.L., and Lipshitz, H.D. 1998. RNA localization in development. Annu. Rev. Biochem. 67: 335–394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
