A central resource for accurate allele frequency estimation from pooled DNA genotyped on DNA microarrays
- PMID: 15701753
- PMCID: PMC549427
- DOI: 10.1093/nar/gni028
A central resource for accurate allele frequency estimation from pooled DNA genotyped on DNA microarrays
Abstract
Analysing pooled DNA on microarrays is an efficient way to genotype hundreds of individuals for thousands of markers for genome-wide association. Although direct comparison of case and control fluorescence scores is possible, correction for differential hybridization of alleles is important, particularly for rare single nucleotide polymorphisms. Such correction relies on heterozygous fluorescence scores and requires the genotyping of hundreds of individuals to obtain sufficient estimates of the correction factor, completely negating any benefit gained by pooling samples. We explore the effect of differential hybridization on test statistics and provide a solution to this problem in the form of a central resource for the accumulation of heterozygous fluorescence scores, allowing accurate allele frequency estimation at no extra cost.
Figures

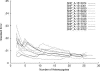
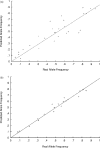
References
-
- Breen G., Harold D., Ralston S., Shaw D., St Clair D. Determining SNP allele frequencies in DNA pools. Biotechniques. 2000;28:464–466. 468, 470. - PubMed
-
- Le Hellard S., Ballereau S.J., Visscher P.M., Torrance H.S., Pinson J., Morris S.W., Thomson M.L., Semple C.A., Muir W.J., Blackwood D.H., Porteous D.J., Evans K.L. SNP genotyping on pooled DNAs: comparison of genotyping technologies and a semi automated method for data storage and analysis. Nucleic Acids Res. 2002;30:e74. - PMC - PubMed
-
- Jawaid A., Bader J.S., Purcell S., Cherry S.S., Sham P.C. Optimal selection strategies for QTL mapping using pooled DNA samples. Eur. J. Hum. Genet. 2002;10:125–132. - PubMed
-
- Norton N., Williams N.M., Williams H.J., Spurlock G., Kirov G., Morris D.W., Hoogendoorn B., Owen M.J., O'Donovan M.C. Universal, robust, highly quantitative SNP allele frequency measurement in DNA pools. Hum. Genet. 2002;110:471–478. - PubMed
-
- Sham P.C., Bader J.S., Craig I., O'Donovan M., Owen M. DNA pooling: a tool for large-scale association studies. Nature Rev. Genet. 2002;3:862–871. - PubMed

