Splicing of human immunodeficiency virus RNA is position-dependent suggesting sequential removal of introns from the 5' end
- PMID: 15701754
- PMCID: PMC549389
- DOI: 10.1093/nar/gki185
Splicing of human immunodeficiency virus RNA is position-dependent suggesting sequential removal of introns from the 5' end
Abstract
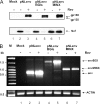
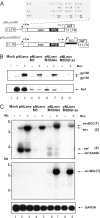
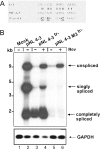
Transcription of the HIV-1 genome yields a single primary transcript, which is alternatively spliced to >30 mRNAs. Productive infection depends on inefficient and regulated splicing and appears to proceed in a tight 5' to 3' order. To analyse whether sequential splicing is mediated by the quality of splice sites or by the position of an intron, we inserted the efficient beta-globin intron (BGI) into the 3' region or 5'UTR of a subgenomic expression vector or an infectious proviral plasmid. RNA analysis revealed splicing of the 3' BGI only if all upstream introns were removed, while splicing of the same intron in the 5'UTR was efficient and independent of further splicing. Furthermore, mutation of the upstream splice signal in the subgenomic vector did not eliminate the inhibition of 3' splicing, although the BGI sequence was the only intron in this case. These results suggest that downstream splicing of HIV-1 RNAs is completely dependent on prior splicing of all upstream intron(s). This hypothesis was supported by the mutation of the major 5' splice site in the HIV-1 genome, which completely abolished all splicing. It appears likely that the tight order of splicing is important for HIV-1 replication, which requires the stable production of intron containing RNAs, while splicing of 3' introns on incompletely spliced RNAs would be likely to render them subject to nonsense-mediated decay.
Figures





References
-
- Coffin J.M., Hughes S.H., Varmus H.E. In: Retroviruses. Coffin J.M., editor. Cold Spring Harbor, NY: 1997. - PubMed
-
- Frankel A.D., Young J.A. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;67:1–25. - PubMed
-
- Pollard V.W., Malim M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. - PubMed
-
- Wodrich H., Krausslich H.G. Nucleocytoplasmic RNA transport in retroviral replication. Results Probl. Cell Differ. 2001;34:197–217. - PubMed
-
- Chang D.D., Sharp P.A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. - PubMed

