Gene discovery and expression profile analysis through sequencing of expressed sequence tags from different developmental stages of the chytridiomycete Blastocladiella emersonii
- PMID: 15701807
- PMCID: PMC549328
- DOI: 10.1128/EC.4.2.455-464.2005
Gene discovery and expression profile analysis through sequencing of expressed sequence tags from different developmental stages of the chytridiomycete Blastocladiella emersonii
Abstract
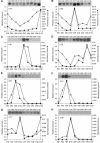
Blastocladiella emersonii is an aquatic fungus of the chytridiomycete class which diverged early from the fungal lineage and is notable for the morphogenetic processes which occur during its life cycle. Its particular taxonomic position makes this fungus an interesting system to be considered when investigating phylogenetic relationships and studying the biology of lower fungi. To contribute to the understanding of the complexity of the B. emersonii genome, we present here a survey of expressed sequence tags (ESTs) from various stages of the fungal development. Nearly 20,000 cDNA clones from 10 different libraries were partially sequenced from their 5' end, yielding 16,984 high-quality ESTs. These ESTs were assembled into 4,873 putative transcripts, of which 48% presented no matches with existing sequences in public databases. As a result of Gene Ontology (GO) project annotation, 1,680 ESTs (35%) were classified into biological processes of the GO structure, with transcription and RNA processing, protein biosynthesis, and transport as prevalent processes. We also report full-length sequences, useful for construction of molecular phylogenies, and several ESTs that showed high similarity with known proteins, some of which were not previously described in fungi. Furthermore, we analyzed the expression profile (digital Northern analysis) of each transcript throughout the life cycle of the fungus using Bayesian statistics. The in silico approach was validated by Northern blot analysis with good agreement between the two methodologies.
Figures



References
-
- Bishop, J. O., J. G. Morton, M. Rosbash, and M. Richardson. 1974. Three abundance classes in HeLa cell messenger RNA. Nature 250:199-204. - PubMed
-
- de Oliveira, J. C., A. C. Borges, M. V. Marques, and S. L. Gomes. 1994. Cloning and characterization of the gene for the catalytic subunit of cAMP-dependent protein kinase in the aquatic fungus Blastocladiella emersonii. Eur. J. Biochem. 219:555-562. - PubMed
-
- de Souza, F. S., and S. L. Gomes. 1998. A P-type ATPase from the aquatic fungus Blastocladiella emersonii similar to animal Na, K-ATPases. Biochim. Biophys. Acta 1383:183-187. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

