Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex
- PMID: 15703211
- PMCID: PMC1087243
- DOI: 10.1091/mbc.e04-10-0857
Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex
Abstract
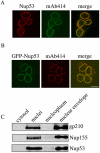
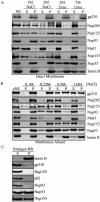
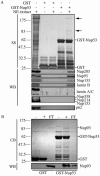
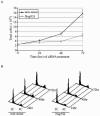
The nuclear pore complex (NPC) is an evolutionarily conserved structure that mediates exchange of macromolecules across the nuclear envelope (NE). It is comprised of approximately 30 proteins termed nucleoporins that are each present in multiple copies. We have investigated the function of the human nucleoporin Nup53, the ortholog of Saccharomyces cerevisiae Nup53p. Both cell fractionation and in vitro binding data suggest that Nup53 is tightly associated with the NE membrane and the lamina where it interacts with lamin B. We have also shown that Nup53 is capable of physically interacting with a group of nucleoporins including Nup93, Nup155, and Nup205. Consistent with this observation, depletion of Nup53 using small interfering RNAs causes a decrease in the cellular levels of these nucleoporins as well as the spindle checkpoint protein Mad1, likely due to destabilization of Nup53-containing complexes. The cellular depletion of this group of nucleoporins, induced by depleting either Nup53 or Nup93, severely alters nuclear morphology producing phenotypes similar to that previously observed in cells depleted of lamin A and Mad1. On basis of these data, we propose a model in which Nup53 is positioned near the pore membrane and the lamina where it anchors an NPC subcomplex containing Nup93, Nup155, and Nup205.
Figures


Similar articles
-
Nup53 is required for nuclear envelope and nuclear pore complex assembly.Mol Biol Cell. 2008 Apr;19(4):1753-62. doi: 10.1091/mbc.e07-08-0820. Epub 2008 Feb 6. Mol Biol Cell. 2008. PMID: 18256286 Free PMC article.
-
A self-inhibitory interaction within Nup155 and membrane binding are required for nuclear pore complex formation.J Cell Sci. 2018 Jan 4;131(1):jcs208538. doi: 10.1242/jcs.208538. J Cell Sci. 2018. PMID: 29150488
-
The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells.Mol Cell. 2006 Apr 7;22(1):93-103. doi: 10.1016/j.molcel.2006.02.015. Mol Cell. 2006. PMID: 16600873
-
Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly.Cells. 2021 Dec 20;10(12):3601. doi: 10.3390/cells10123601. Cells. 2021. PMID: 34944108 Free PMC article. Review.
-
Autoantigens of the nuclear pore complex.J Mol Med (Berl). 2004 Jul;82(7):423-33. doi: 10.1007/s00109-004-0554-z. Epub 2004 Jun 3. J Mol Med (Berl). 2004. PMID: 15175862 Review.
Cited by
-
Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly.EMBO J. 2012 Oct 17;31(20):4072-84. doi: 10.1038/emboj.2012.256. Epub 2012 Sep 7. EMBO J. 2012. PMID: 22960634 Free PMC article.
-
Biallelic Mutations in Nuclear Pore Complex Subunit NUP107 Cause Early-Childhood-Onset Steroid-Resistant Nephrotic Syndrome.Am J Hum Genet. 2015 Oct 1;97(4):555-66. doi: 10.1016/j.ajhg.2015.08.013. Epub 2015 Sep 24. Am J Hum Genet. 2015. PMID: 26411495 Free PMC article.
-
The nuclear envelope: form and reformation.Curr Opin Cell Biol. 2006 Feb;18(1):108-16. doi: 10.1016/j.ceb.2005.12.004. Epub 2005 Dec 20. Curr Opin Cell Biol. 2006. PMID: 16364623 Free PMC article. Review.
-
Biallelic loss of function NEK3 mutations deacetylate α-tubulin and downregulate NUP205 that predispose individuals to cilia-related abnormal cardiac left-right patterning.Cell Death Dis. 2020 Nov 23;11(11):1005. doi: 10.1038/s41419-020-03214-1. Cell Death Dis. 2020. PMID: 33230144 Free PMC article.
-
A single-molecule localization microscopy method for tissues reveals nonrandom nuclear pore distribution in Drosophila.J Cell Sci. 2021 Dec 15;134(24):jcs259570. doi: 10.1242/jcs.259570. Epub 2021 Dec 16. J Cell Sci. 2021. PMID: 34806753 Free PMC article.
References
-
- Aitchison, J. D., Rout, M. P., Marelli, M., Blobel, G., and Wozniak, R. W. (1995b). Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131, 1133-1148. - PMC - PubMed
-
- Allen, N. P., Patel, S. S., Huang, L., Chalkley, R. J., Burlingame, A., Lutzmann, M., Hurt, E. C., and Rexach, M. (2002). Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell Proteomics 1, 930-946. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

