Presenilin-1-dependent transcriptome changes
- PMID: 15703411
- PMCID: PMC6726008
- DOI: 10.1523/JNEUROSCI.4145-04.2005
Presenilin-1-dependent transcriptome changes
Abstract
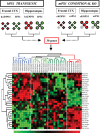
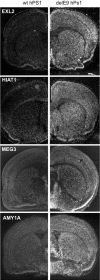
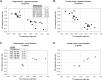
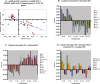
Familial forms of Alzheimer's disease (FADs) are caused by the expression of mutant presenilin 1 (PS1) or presenilin 2. Using DNA microarrays, we explored the brain transcription profiles of mice with conditional knock-out of PS1 (cKO PS1) in the forebrain. In parallel, we performed a transcription profiling of the hippocampus and frontal cortex of the FAD-linked DeltaE9 mutant transgenic (TG) mice and matched controls [TG mice expressing wild-type human PS1 (hPS1)]. When the TG and cKO datasets were cross-compared, the majority of the 30 common expression alterations were in opposite direction, suggesting that the FAD-linked PS1 variant produces transcriptome changes primarily by gain of aberrant function. Our microarray studies also revealed an unanticipated inverse correlation of transcript levels between the brains of mice that coexpress DeltaE9 hPS1+ amyloid precursor protein (APP)695 Swe and DeltaE9 hPS1 single transgenic mice. The opposite directionality of these changes in transcript levels must be a function of APP and/or APP derivatives.
Figures

References
-
- Anderson AJ, Cummings BJ, Cotman CW (1994) Increased immunoreactivity for Jun- and Fos-related proteins in Alzheimer's disease: association with pathology. Exp Neurol 125: 286-295. - PubMed
-
- Ball CA, Sherlock G, Parkinson H, Rocca-Sera P, Brooksbank C, Causton HC, Cavalieri D, Gaasterland T, Hingamp P, Holstege F, Ringwald M, Spellman P, Stoeckert Jr CJ, Stewart JE, Taylor R, Brazma A, Quackenbush J (2002) Standards for microarray data. Science 298: 539. - PubMed
-
- Borchelt DR, Thinakaran G, Eckman CB, Lee MK, Davenport F, Ratovitsky T, Prada CM, Kim G, Seekins S, Yager D, Slunt HH, Wang R, Seeger M, Levey AI, Gandy SE, Copeland NG, Jenkins NA, Price DL, Younkin SG, Sisodia SS (1996) Familial Alzheimer's disease-linked presenilin 1 variants elevate Abeta1-42/1-40 ratio in vitro and in vivo Neuron 17: 1005-1013. - PubMed
-
- Borchelt DR, Ratovitski T, van Lare J, Lee MK, Gonzales V, Jenkins NA, Copeland NG, Price DL, Sisodia SS (1997) Accelerated amyloid deposition in the brains of transgenic mice coexpressing mutant presenilin 1 and amyloid precursor proteins. Neuron 19: 939-945. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
