Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association
- PMID: 15703437
- PMCID: PMC1370736
- DOI: 10.1261/rna.7215305
Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association
Abstract
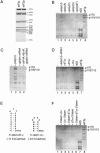
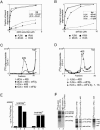
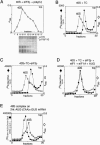
The multisubunit eukaryotic initiation factor (eIF) 3 plays various roles in translation initiation that all involve interaction with 40S ribosomal subunits. eIF3 can be purified in two forms: with or without the loosely associated eIF3j subunit (eIF3j+ and eIF3j-, respectively). Although unlike eIF3j+, eIF3j- does not bind 40S subunits stably enough to withstand sucrose density gradient centrifugation, we found that in addition to the known stabilization of the eIF3/40S subunit interaction by the eIF2*GTP*Met-tRNA(i)Met ternary complex, eIF3j-/40S subunit complexes were also stabilized by single-stranded RNA or DNA cofactors that were at least 25 nt long and could be flanked by stable hairpins. Of all homopolymers, oligo(rU), oligo(dT), and oligo(dC) stimulated the eIF3/40S subunit interaction, whereas oligo(rA), oligo(rG), oligo(rC), oligo(dA), and oligo(dG) did not. Oligo(U) or oligo(dT) sequences interspersed by other bases also promoted this interaction. The ability of oligonucleotides to stimulate eIF3/40S subunit association correlated with their ability to bind to the 40S subunit, most likely to its mRNA-binding cleft. Although eIF3j+ could bind directly to 40S subunits, neither eIF3j- nor eIF3j+ alone was able to dissociate 80S ribosomes or protect 40S and 60S subunits from reassociation. Significantly, the dissociation/anti-association activities of both forms of eIF3 became apparent in the presence of either eIF2-ternary complexes or any oligonucleotide cofactor that promoted eIF3/40S subunit interaction. Ribosomal dissociation and anti-association activities of eIF3 were strongly enhanced by eIF1. The potential biological role of stimulation of eIF3/40S subunit interaction by an RNA cofactor in the absence of eIF2-ternary complex is discussed.
Figures




Similar articles
-
Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast.Mol Cell Biol. 2006 Feb;26(4):1355-72. doi: 10.1128/MCB.26.4.1355-1372.2006. Mol Cell Biol. 2006. PMID: 16449648 Free PMC article.
-
The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro.J Biol Chem. 2004 Mar 5;279(10):8946-56. doi: 10.1074/jbc.M312745200. Epub 2003 Dec 19. J Biol Chem. 2004. PMID: 14688252
-
eIF3j is located in the decoding center of the human 40S ribosomal subunit.Mol Cell. 2007 Jun 22;26(6):811-9. doi: 10.1016/j.molcel.2007.05.019. Mol Cell. 2007. PMID: 17588516
-
eIF3: a versatile scaffold for translation initiation complexes.Trends Biochem Sci. 2006 Oct;31(10):553-62. doi: 10.1016/j.tibs.2006.08.005. Epub 2006 Aug 22. Trends Biochem Sci. 2006. PMID: 16920360 Review.
-
Molecular mechanisms of translation initiation in eukaryotes.Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7029-36. doi: 10.1073/pnas.111145798. Proc Natl Acad Sci U S A. 2001. PMID: 11416183 Free PMC article. Review.
Cited by
-
eIF5 and eIF5B together stimulate 48S initiation complex formation during ribosomal scanning.Nucleic Acids Res. 2014 Oct 29;42(19):12052-69. doi: 10.1093/nar/gku877. Epub 2014 Sep 26. Nucleic Acids Res. 2014. PMID: 25260592 Free PMC article.
-
The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection.J Mol Biol. 2010 Mar 5;396(4):1097-116. doi: 10.1016/j.jmb.2009.12.047. Epub 2010 Jan 11. J Mol Biol. 2010. PMID: 20060839 Free PMC article.
-
Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex.Elife. 2016 Oct 26;5:e20934. doi: 10.7554/eLife.20934. Elife. 2016. PMID: 27782884 Free PMC article.
-
Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP.Genes Dev. 2004 Dec 15;18(24):3078-93. doi: 10.1101/gad.1255704. Genes Dev. 2004. PMID: 15601822 Free PMC article.
-
The eIF3 complex of Trypanosoma brucei: composition conservation does not imply the conservation of structural assembly and subunits function.RNA. 2017 Mar;23(3):333-345. doi: 10.1261/rna.058651.116. Epub 2016 Dec 8. RNA. 2017. PMID: 27932584 Free PMC article.
References
-
- Arnott, S., Chandrasekaran, R., and Leslie, A.G. 1976. Structure of the single-stranded polyribonucleotide polycytidylic acid. J. Mol. Biol. 106: 735–748. - PubMed
-
- Asano, K., Kinzy, T.G., Merrick, W.C., and Hershey, J.W.B. 1997. Conservation and diversity of eukaryotic translation initiation factor eIF3. J. Biol. Chem. 272: 1101–1109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
