Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles
- PMID: 15703440
- PMCID: PMC1370728
- DOI: 10.1261/rna.7234205
Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles
Abstract
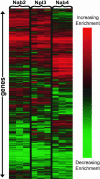
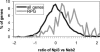
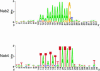
Nab2, Npl3, and Nab4/Hrp1 are essential RNA binding proteins of the shuttling hnRNP class that are required for the efficient export of mRNA. To characterize the in vivo transcript specificity of these proteins, we identified their mRNA binding partners using a microarray-based assay. Each of the three proteins was coimmunoprecipitated with many different mRNA transcripts. Interestingly, each protein exhibits preferential associations with a distinct set of mRNAs. Notably, some of these appear to denote specific functional classes. For example, the ribosomal protein mRNAs and other highly expressed transcripts significantly favor association with Npl3 over Nab2, and Nab4/Hrp1 is strongly enriched with transcripts required for amino acid metabolism. Significantly, nab4 mutants showed a striking, desensitized growth phenotype when exposed to amino acid stress conditions suggesting a biological consequence to the associations we observed. Supporting the hypothesis that these proteins display transcript specificity, we identified a unique 7-nucleotide sequence overrepresented in the transcripts highly associated with Nab2 and Nab4/Hrp1 using the REDUCE algorithm. Validating our approach, our bioinformatics analysis correctly identified the known binding site for Nab4/Hrp1. These specialized associations of the hnRNP proteins of Saccharomyces cerevisiae suggest the opportunity to regulate the processing of particular transcripts between transcription and translation.
Figures

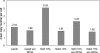
Similar articles
-
The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay.Mol Cell. 2000 Mar;5(3):489-99. doi: 10.1016/s1097-2765(00)80443-8. Mol Cell. 2000. PMID: 10882134
-
Alternative 3' pre-mRNA processing in Saccharomyces cerevisiae is modulated by Nab4/Hrp1 in vivo.PLoS Biol. 2007 Jan;5(1):e6. doi: 10.1371/journal.pbio.0050006. PLoS Biol. 2007. PMID: 17194212 Free PMC article.
-
Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export.J Biol Chem. 2008 Oct 3;283(40):27130-43. doi: 10.1074/jbc.M803649200. Epub 2008 Aug 5. J Biol Chem. 2008. PMID: 18682389 Free PMC article.
-
Structure-function relationships in the Nab2 polyadenosine-RNA binding Zn finger protein family.Protein Sci. 2019 Mar;28(3):513-523. doi: 10.1002/pro.3565. Epub 2019 Jan 16. Protein Sci. 2019. PMID: 30578643 Free PMC article. Review.
-
[Nuclear transport of hnRNP and mRNA].Tanpakushitsu Kakusan Koso. 2000 Oct;45(14):2378-87. Tanpakushitsu Kakusan Koso. 2000. PMID: 11051839 Review. Japanese. No abstract available.
Cited by
-
Yeast hnRNP-related proteins contribute to the maintenance of telomeres.Biochem Biophys Res Commun. 2012 Sep 14;426(1):12-7. doi: 10.1016/j.bbrc.2012.07.144. Epub 2012 Aug 10. Biochem Biophys Res Commun. 2012. PMID: 22902537 Free PMC article.
-
mRNA nuclear export at a glance.J Cell Sci. 2009 Jun 15;122(Pt 12):1933-7. doi: 10.1242/jcs.041236. J Cell Sci. 2009. PMID: 19494120 Free PMC article. Review. No abstract available.
-
Hydrolase regulates NAD+ metabolites and modulates cellular redox.J Biol Chem. 2009 Apr 24;284(17):11256-66. doi: 10.1074/jbc.M809790200. Epub 2009 Feb 27. J Biol Chem. 2009. PMID: 19251690 Free PMC article.
-
fREDUCE: detection of degenerate regulatory elements using correlation with expression.BMC Bioinformatics. 2007 Oct 17;8:399. doi: 10.1186/1471-2105-8-399. BMC Bioinformatics. 2007. PMID: 17941998 Free PMC article.
-
The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability.Genes Dev. 2013 Nov 15;27(22):2445-58. doi: 10.1101/gad.229880.113. Genes Dev. 2013. PMID: 24240235 Free PMC article.
References
-
- Bailey, T.L. and Elkan, C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. - PubMed
-
- Blanchette, M., Labourier, E., Green, R.E., Brenner, S.E., and Rio, D.C. 2004. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF(50) in the nuclear export of intronless mRNAs. Mol. Cell 14: 775–786. - PubMed
-
- Brown, V., Jin, P., Ceman, S., Darnell, J.C., O’Donnell, W.T., Tenenbaum, S.A., Jin, X., Feng, Y., Wilkinson, K.D., Keene, J.D., et al. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107: 477–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
