Ribozyme motif structure mapped using random recombination and selection
- PMID: 15703441
- PMCID: PMC1370730
- DOI: 10.1261/rna.7238705
Ribozyme motif structure mapped using random recombination and selection
Abstract
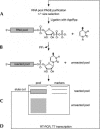
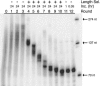
Isolating the core functional elements of an RNA is normally performed during the characterization of a new RNA in order to simplify further biochemical analysis. The removal of extraneous sequence is challenging and can lead to biases that result from the incomplete sampling of deletion variants. An impartial solution to this problem is to construct a library containing a large number of deletion constructs and to select functional RNA isolates that are at least as efficient as their full-length progenitors. Here, we use nonhomologous recombination and selection to isolate the catalytic core of a pyrimidine nucleotide synthase ribozyme. A variable-length pool of approximately 10(8) recombinant molecules that included deletions, inversions, and translocations of a 271-nucleotide-long ribozyme isolate was constructed by digesting and randomly religating its DNA genome. In vitro selection for functional ribozymes was then performed in a size-dependent and a size-independent manner. The final pools had nearly equivalent catalytic rates even though their length distributions were completely different, indicating that a diverse range of deletion constructs were functionally active. Four short sequence islands, requiring as little as 81 nt of sequence, were found within all of the truncated ribozymes and could be folded into a secondary structure consisting of three helix-loops. Our findings suggest that nonhomologous recombination is a highly efficient way to isolate a ribozyme's core motif and could prove to be a useful method for evolving new ribozyme functions from pre-existing sequences in a manner that may have played an important role early in evolution.
Figures




Similar articles
-
Combinatorial minimization and secondary structure determination of a nucleotide synthase ribozyme.RNA. 2003 Oct;9(10):1208-20. doi: 10.1261/rna.5500603. RNA. 2003. PMID: 13130135 Free PMC article.
-
In vitro evolution suggests multiple origins for the hammerhead ribozyme.Nature. 2001 Nov 1;414(6859):82-4. doi: 10.1038/35102081. Nature. 2001. PMID: 11689947
-
New catalytic structures from an existing ribozyme.Nat Struct Mol Biol. 2005 Nov;12(11):994-1000. doi: 10.1038/nsmb1003. Nat Struct Mol Biol. 2005. PMID: 16228005
-
The many faces of the hairpin ribozyme: structural and functional variants of a small catalytic RNA.IUBMB Life. 2012 Jan;64(1):36-47. doi: 10.1002/iub.575. Epub 2011 Nov 30. IUBMB Life. 2012. PMID: 22131309 Review.
-
Evolutionary origins and directed evolution of RNA.Int J Biochem Cell Biol. 2009 Feb;41(2):254-65. doi: 10.1016/j.biocel.2008.08.015. Epub 2008 Aug 19. Int J Biochem Cell Biol. 2009. PMID: 18775793 Review.
Cited by
-
Modular evolution and increase of functional complexity in replicating RNA molecules.RNA. 2007 Jan;13(1):97-107. doi: 10.1261/rna.203006. Epub 2006 Nov 14. RNA. 2007. PMID: 17105993 Free PMC article.
-
Synthetic shuffling and in vitro selection reveal the rugged adaptive fitness landscape of a kinase ribozyme.RNA. 2013 Aug;19(8):1116-28. doi: 10.1261/rna.037572.112. Epub 2013 Jun 24. RNA. 2013. PMID: 23798664 Free PMC article.
-
Ebbie: automated analysis and storage of small RNA cloning data using a dynamic web server.BMC Bioinformatics. 2006 Apr 3;7:185. doi: 10.1186/1471-2105-7-185. BMC Bioinformatics. 2006. PMID: 16584563 Free PMC article.
-
Recombination during in vitro evolution.J Mol Evol. 2005 Aug;61(2):245-52. doi: 10.1007/s00239-004-0373-4. Epub 2005 Jun 30. J Mol Evol. 2005. PMID: 16007485
-
Functional information and the emergence of biocomplexity.Proc Natl Acad Sci U S A. 2007 May 15;104 Suppl 1(Suppl 1):8574-81. doi: 10.1073/pnas.0701744104. Epub 2007 May 9. Proc Natl Acad Sci U S A. 2007. PMID: 17494745 Free PMC article.
References
-
- Campbell, V.W. and Jackson, D.A. 1980. The effect of divalent cations on the mode of action of DNase I. The initial reaction products produced from covalently closed circular DNA. J. Biol. Chem. 255: 3726–3735. - PubMed
-
- Ekland, E.H., Szostak, J.W., and Bartel, D.P. 1995. Structurally complex and highly-active RNA ligases derived from random RNA sequences. Science 269: 364–370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
