Processing bodies require RNA for assembly and contain nontranslating mRNAs
- PMID: 15703442
- PMCID: PMC1370727
- DOI: 10.1261/rna.7258505
Processing bodies require RNA for assembly and contain nontranslating mRNAs
Abstract
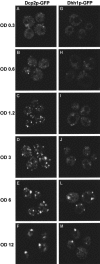
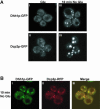
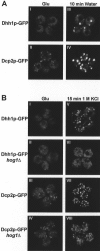
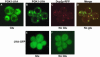
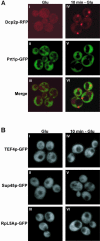
Recent experiments have defined cytoplasmic foci, referred to as processing bodies (P-bodies), wherein mRNA decay factors are concentrated and where mRNA decay can occur. However, the physical nature of P-bodies, their relationship to translation, and possible roles of P-bodies in cellular responses remain unclear. We describe four properties of yeast P-bodies that indicate that P-bodies are dynamic structures that contain nontranslating mRNAs and function during cellular responses to stress. First, in vivo and in vitro analysis indicates that P-bodies are dependent on RNA for their formation. Second, the number and size of P-bodies vary in response to glucose deprivation, osmotic stress, exposure to ultraviolet light, and the stage of cell growth. Third, P-bodies vary with the status of the cellular translation machinery. Inhibition of translation initiation by mutations, or cellular stress, results in increased P-bodies. In contrast, inhibition of translation elongation, thereby trapping the mRNA in polysomes, leads to dissociation of P-bodies. Fourth, multiple translation factors and ribosomal proteins are lacking from P-bodies. These results suggest additional biological roles of P-bodies in addition to being sites of mRNA degradation.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
