Allele-specific assay reveals functional variation in the chalcone synthase promoter of Arabidopsis thaliana that is compatible with neutral evolution
- PMID: 15705952
- PMCID: PMC1069691
- DOI: 10.1105/tpc.104.027839
Allele-specific assay reveals functional variation in the chalcone synthase promoter of Arabidopsis thaliana that is compatible with neutral evolution
Abstract
Promoters are thought to play a major role in adaptive evolution, yet little is known about the regulatory diversity within species, where microevolutionary processes take place. To investigate the potential for evolutionary change in the promoter of a gene, we examined nucleotide and functional variation of the Chalcone Synthase (CHS) cis-regulatory region in Arabidopsis thaliana. CHS is the branch point enzyme of a biosynthetic pathway that leads to the production of secondary metabolites influencing the interaction between the plant and its environment. We found that nucleotide diversity in the intergenic region encompassing the CHS promoter (pi=0.003) is compatible with neutral expectations. To quantify functional variation specifically as a result of cis-regulation of CHS mRNA levels, we developed an assay using F1 individuals in which distinct promoter alleles are compared within a common trans-regulatory background. We examined functional cis-regulatory variation in response to different stimuli representing a variety of CHS transcriptional environments (dark, light, and insect feeding). We observed extensive functional variation, some of which appeared to be independent of the trans-regulatory background. Comparison of functional and nucleotide diversity suggested a candidate point mutation that may explain cis-regulatory differences in light response. Our results indicate that functional changes in promoters can arise from a few mutations, pointing to promoter regions as a fundamental determinant of functional genetic variation.
Figures
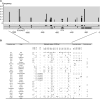

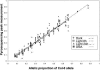
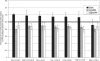
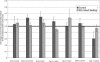
References
-
- Ahmadian, A., Gharizadeh, B., Gustafsson, A.C., Sterky, F., Nyren, P., Uhlen, M., and Lundeberg, J. (2000). Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem. 280, 103–110. - PubMed
-
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. (2003). Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299, 1391–1394. - PubMed
-
- Bray, N.J., Buckland, P.R., Owen, M.J., and O'Donovan, M.C. (2003). Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 113, 149–153. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

