The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development
- PMID: 15705953
- PMCID: PMC1069696
- DOI: 10.1105/tpc.104.027714
The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development
Abstract
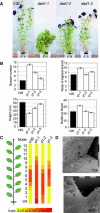
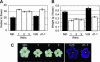
Carotenoids and carotenoid cleavage products play an important and integral role in plant development. The Decreased apical dominance1 (Dad1)/PhCCD8 gene of petunia (Petunia hybrida) encodes a hypothetical carotenoid cleavage dioxygenase (CCD) and ortholog of the MORE AXILLARY GROWTH4 (MAX4)/AtCCD8 gene. The dad1-1 mutant allele was inactivated by insertion of an unusual transposon (Dad-one transposon), and the dad1-3 allele is a revertant allele of dad1-1. Consistent with its role in producing a graft-transmissible compound that can alter branching, the Dad1/PhCCD8 gene is expressed in root and shoot tissue. This expression is upregulated in the stems of the dad1-1, dad2, and dad3 increased branching mutants, indicating feedback regulation of the gene in this tissue. However, this feedback regulation does not affect the root expression of Dad1/PhCCD8. Overexpression of Dad1/PhCCD8 in the dad1-1 mutant complemented the mutant phenotype, and RNA interference in the wild type resulted in an increased branching phenotype. Other differences in phenotype associated with the loss of Dad1/PhCCD8 function included altered timing of axillary meristem development, delayed leaf senescence, smaller flowers, reduced internode length, and reduced root growth. These data indicate that the substrate(s) and/or product(s) of the Dad1/PhCCD8 enzyme are mobile signal molecules with diverse roles in plant development.
Figures



References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Beers, E., Moreno, T., and Callis, J. (1992). Subcellular localization of ubiquitin and ubiquitinated proteins in Arabidopsis thaliana. J. Biol. Chem. 267, 15432–15439. - PubMed
-
- Bell, A.D. (1991). Plant Form: An Illustrated Guide to Flowering Plant Morphology. (Oxford: Oxford University Press).
-
- Beveridge, C.A. (2000). Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul. 32, 193–203.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

