An initial blueprint for myogenic differentiation
- PMID: 15706034
- PMCID: PMC551576
- DOI: 10.1101/gad.1281105
An initial blueprint for myogenic differentiation
Abstract
We have combined genome-wide transcription factor binding and expression profiling to assemble a regulatory network controlling the myogenic differentiation program in mammalian cells. We identified a cadre of overlapping and distinct targets of the key myogenic regulatory factors (MRFs)--MyoD and myogenin--and Myocyte Enhancer Factor 2 (MEF2). We discovered that MRFs and MEF2 regulate a remarkably extensive array of transcription factor genes that propagate and amplify the signals initiated by MRFs. We found that MRFs play an unexpectedly wide-ranging role in directing the assembly and usage of the neuromuscular junction. Interestingly, these factors also prepare myoblasts to respond to diverse types of stress. Computational analyses identified novel combinations of factors that, depending on the differentiation state, might collaborate with MRFs. Our studies suggest unanticipated biological insights into muscle development and highlight new directions for further studies of genes involved in muscle repair and responses to stress and damage.
Figures

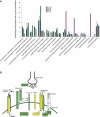
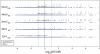
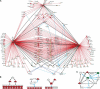
References
-
- Alon U. 2003. Biological networks: The tinkerer as an engineer. Science 301: 1866-1867. - PubMed
-
- Benezra R., Davis, R.L., Lockshon, D., Turner, D.L., and Weintraub, H. 1990. The protein Id: A negative regulator of helixloop-helix DNA binding proteins. Cell 61: 49-59. - PubMed
-
- Bergstrom D.A., Penn, B.H., Strand, A., Perry, R.L., Rudnicki, M.A., and Tapscott, S.J. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell 9: 587-600. - PubMed
-
- Buckingham M. 2001. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11: 440-448. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
