Ontological visualization of protein-protein interactions
- PMID: 15707487
- PMCID: PMC550656
- DOI: 10.1186/1471-2105-6-29
Ontological visualization of protein-protein interactions
Abstract
Background: Cellular processes require the interaction of many proteins across several cellular compartments. Determining the collective network of such interactions is an important aspect of understanding the role and regulation of individual proteins. The Gene Ontology (GO) is used by model organism databases and other bioinformatics resources to provide functional annotation of proteins. The annotation process provides a mechanism to document the binding of one protein with another. We have constructed protein interaction networks for mouse proteins utilizing the information encoded in the GO annotations. The work reported here presents a methodology for integrating and visualizing information on protein-protein interactions.
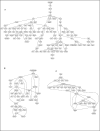
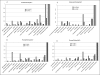
Results: GO annotation at Mouse Genome Informatics (MGI) captures 1318 curated, documented interactions. These include 129 binary interactions and 125 interaction involving three or more gene products. Three networks involve over 30 partners, the largest involving 109 proteins. Several tools are available at MGI to visualize and analyze these data.
Conclusions: Curators at the MGI database annotate protein-protein interaction data from experimental reports from the literature. Integration of these data with the other types of data curated at MGI places protein binding data into the larger context of mouse biology and facilitates the generation of new biological hypotheses based on physical interactions among gene products.
Figures




Similar articles
-
A procedure for assessing GO annotation consistency.Bioinformatics. 2005 Jun;21 Suppl 1:i136-43. doi: 10.1093/bioinformatics/bti1019. Bioinformatics. 2005. PMID: 15961450
-
AVID: an integrative framework for discovering functional relationships among proteins.BMC Bioinformatics. 2005 Jun 1;6:136. doi: 10.1186/1471-2105-6-136. BMC Bioinformatics. 2005. PMID: 15929793 Free PMC article.
-
Searching the Mouse Genome Informatics (MGI) resources for information on mouse biology from genotype to phenotype.Curr Protoc Bioinformatics. 2004 May;Chapter 1:Unit 1.7. doi: 10.1002/0471250953.bi0107s05. Curr Protoc Bioinformatics. 2004. PMID: 18428715
-
Ontology annotation: mapping genomic regions to biological function.Curr Opin Chem Biol. 2007 Feb;11(1):4-11. doi: 10.1016/j.cbpa.2006.11.039. Epub 2007 Jan 5. Curr Opin Chem Biol. 2007. PMID: 17208035 Review.
-
Mouse Genome Informatics (MGI) Is the International Resource for Information on the Laboratory Mouse.Methods Mol Biol. 2018;1757:141-161. doi: 10.1007/978-1-4939-7737-6_7. Methods Mol Biol. 2018. PMID: 29761459 Review.
Cited by
-
Systematic variation in mRNA 3'-processing signals during mouse spermatogenesis.Nucleic Acids Res. 2007;35(1):234-46. doi: 10.1093/nar/gkl919. Epub 2006 Dec 8. Nucleic Acids Res. 2007. PMID: 17158511 Free PMC article.
-
Biomedical discovery acceleration, with applications to craniofacial development.PLoS Comput Biol. 2009 Mar;5(3):e1000215. doi: 10.1371/journal.pcbi.1000215. Epub 2009 Mar 27. PLoS Comput Biol. 2009. PMID: 19325874 Free PMC article.
-
Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae.J Biol. 2006;5(4):11. doi: 10.1186/jbiol36. Epub 2006 Jun 8. J Biol. 2006. PMID: 16762047 Free PMC article.
-
The BioGRID Interaction Database: 2011 update.Nucleic Acids Res. 2011 Jan;39(Database issue):D698-704. doi: 10.1093/nar/gkq1116. Epub 2010 Nov 11. Nucleic Acids Res. 2011. PMID: 21071413 Free PMC article.
-
GeneLibrarian: an effective gene-information summarization and visualization system.BMC Bioinformatics. 2006 Aug 29;7:392. doi: 10.1186/1471-2105-7-392. BMC Bioinformatics. 2006. PMID: 16939640 Free PMC article.
References
-
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

