Evolution of the relaxin-like peptide family
- PMID: 15707501
- PMCID: PMC551602
- DOI: 10.1186/1471-2148-5-14
Evolution of the relaxin-like peptide family
Abstract
Background: The relaxin-like peptide family belongs in the insulin superfamily and consists of 7 peptides of high structural but low sequence similarity; relaxin-1, 2 and 3, and the insulin-like (INSL) peptides, INSL3, INSL4, INSL5 and INSL6. The functions of relaxin-3, INSL4, INSL5, INSL6 remain uncharacterised. The evolution of this family has been contentious; high sequence variability is seen between closely related species, while distantly related species show high similarity; an invertebrate relaxin sequence has been reported, while a relaxin gene has not been found in the avian and ruminant lineages.
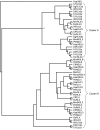
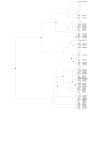
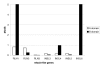
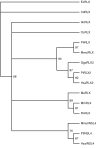
Results: Sequence similarity searches of genomic and EST data identified homologs of relaxin-like peptides in mammals, and non-mammalian vertebrates such as fish. Phylogenetic analysis was used to resolve the evolution of the family. Searches were unable to identify an invertebrate relaxin-like peptide. The published relaxin cDNA sequence in the tunicate, Ciona intestinalis was not present in the completed C. intestinalis genome. The newly discovered relaxin-3 is likely to be the ancestral relaxin. Multiple relaxin-3-like sequences are present in fugu fish (Takifugu rubripes) and zebrafish (Danio rerio), but these appear to be specific to the fish lineage. Possible relaxin-1 and INSL5 homologs were also identified in fish and frog species, placing their emergence prior to mammalia, earlier than previously believed. Furthermore, estimates of synonymous and nonsynonymous substitution rates (dN/dS) suggest that the emergence of relaxin-1, INSL4 and INSL6 during mammalia was driven by positive Darwinian selection, hence these peptides are likely to have novel and in the case of relaxin-1, which is still under positive selection in humans and the great apes, possibly still evolving functions. In contrast, relaxin-3 is constrained by strong purifying selection, demonstrating it must have a highly conserved function, supporting its hypothesized important neuropeptide role.
Conclusions: We present a phylogeny describing the evolutionary history of the relaxin-like peptide family and show that positive selection has driven the evolution of the most recent members of the family.
Figures

Similar articles
-
Evolution of the relaxin-like peptide family: from neuropeptide to reproduction.Ann N Y Acad Sci. 2005 May;1041:530-3. doi: 10.1196/annals.1282.079. Ann N Y Acad Sci. 2005. PMID: 15956756
-
Relaxin gene family in teleosts: phylogeny, syntenic mapping, selective constraint, and expression analysis.BMC Evol Biol. 2009 Dec 16;9:293. doi: 10.1186/1471-2148-9-293. BMC Evol Biol. 2009. PMID: 20015397 Free PMC article.
-
The evolution of the relaxin peptide family and their receptors.Adv Exp Med Biol. 2007;612:1-13. doi: 10.1007/978-0-387-74672-2_1. Adv Exp Med Biol. 2007. PMID: 18161477 Review.
-
Gene turnover and differential retention in the relaxin/insulin-like gene family in primates.Mol Phylogenet Evol. 2012 Jun;63(3):768-76. doi: 10.1016/j.ympev.2012.02.011. Epub 2012 Mar 1. Mol Phylogenet Evol. 2012. PMID: 22405815
-
The relaxin family peptide receptors and their ligands: new developments and paradigms in the evolution from jawless fish to mammals.Gen Comp Endocrinol. 2014 Dec 1;209:93-105. doi: 10.1016/j.ygcen.2014.07.014. Epub 2014 Jul 29. Gen Comp Endocrinol. 2014. PMID: 25079565 Review.
Cited by
-
Identification of prohormones and pituitary neuropeptides in the African cichlid, Astatotilapia burtoni.BMC Genomics. 2016 Aug 19;17(1):660. doi: 10.1186/s12864-016-2914-9. BMC Genomics. 2016. PMID: 27543050 Free PMC article.
-
Elucidation of relaxin-3 binding interactions in the extracellular loops of RXFP3.Front Endocrinol (Lausanne). 2013 Feb 22;4:13. doi: 10.3389/fendo.2013.00013. eCollection 2013. Front Endocrinol (Lausanne). 2013. PMID: 23440673 Free PMC article.
-
Distinct but overlapping binding sites of agonist and antagonist at the relaxin family peptide 3 (RXFP3) receptor.J Biol Chem. 2018 Oct 12;293(41):15777-15789. doi: 10.1074/jbc.RA118.002645. Epub 2018 Aug 21. J Biol Chem. 2018. PMID: 30131340 Free PMC article.
-
The relaxin receptor (RXFP1) utilizes hydrophobic moieties on a signaling surface of its N-terminal low density lipoprotein class A module to mediate receptor activation.J Biol Chem. 2013 Sep 27;288(39):28138-51. doi: 10.1074/jbc.M113.499640. Epub 2013 Aug 7. J Biol Chem. 2013. PMID: 23926099 Free PMC article.
-
Role of sphingosine kinase/S1P axis in ECM remodeling of cardiac cells elicited by relaxin.Mol Endocrinol. 2015 Jan;29(1):53-67. doi: 10.1210/me.2014-1201. Mol Endocrinol. 2015. PMID: 25415609 Free PMC article.
References
-
- Bathgate RA, Samuel CS, Burazin TC, Layfield S, Claasz AA, Reytomas IG, Dawson NF, Zhao C, Bond C, Summers RJ, Parry LJ, Wade JD, Tregear GW. Human relaxin gene 3 (H3) and the equivalent mouse relaxin (M3) gene. Novel members of the relaxin peptide family. Journal of Biological Chemistry. 2002;277:1148–1157. doi: 10.1074/jbc.M107882200. - DOI - PubMed
-
- Hansell DJ, Bryant-Greenwood GD, Greenwood FC. Expression of the human relaxin H1 gene in the decidua, trophoblast, and prostate. Journal of Clinical Endocrinology and Metabolism. 1991;72:899–904. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

