Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: modes of ligand binding and dimerization
- PMID: 15708973
- PMCID: PMC549502
- DOI: 10.1073/pnas.0409913102
Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: modes of ligand binding and dimerization
Abstract
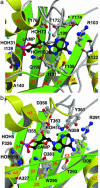
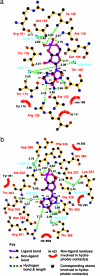
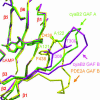
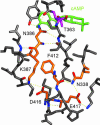
In several species, GAF domains, which are widely expressed small-molecule-binding domains that regulate enzyme activity, are known to bind cyclic nucleotides. However, the molecular mechanism by which cyclic nucleotide binding affects enzyme activity is not known for any GAF domain. In the cyanobacterium, Anabaena, the cyaB1 and cyaB2 genes encode adenylyl cyclases that are stimulated by binding of cAMP to their N-terminal GAF domains. Replacement of the tandem GAF-A/B domains in cyaB1 with the mammalian phosphodiesterase 2A GAF-A/B tandem domains allows regulation of the chimeric protein by cGMP, suggesting a highly conserved mechanism of activation. Here, we describe the 1.9-A crystal structure of the tandem GAF-A/B domains of cyaB2 with bound cAMP and compare it to the previously reported structure of the PDE2A GAF-A/B. Unexpectedly, the cyaB2 GAF-A/B dimer is antiparallel, unlike the parallel dimer of PDE2A. Moreover, there is clear electron density for cAMP in both GAF-A and -B, whereas in PDE2A, cGMP is found only in GAF-B. Phosphate and ribose group contacts are similar to those in PDE2A. However, the purine-binding pockets appear very different from that in PDE2A GAF-B. Differences in the beta2-beta3 loop suggest that this loop confers much of the ligand specificity in this and perhaps in many other GAF domains. Finally, a conserved asparagine appears to be a new addition to the signature NKFDE motif, and a mechanism for this motif to stabilize the cNMP-binding pocket is proposed.
Figures


Similar articles
-
The cyanobacterial tandem GAF domains from the cyaB2 adenylyl cyclase signal via both cAMP-binding sites.Proc Natl Acad Sci U S A. 2005 Feb 22;102(8):3088-92. doi: 10.1073/pnas.0409917102. Epub 2005 Feb 11. Proc Natl Acad Sci U S A. 2005. PMID: 15708972 Free PMC article.
-
Crystal structure of the GAF-B domain from human phosphodiesterase 10A complexed with its ligand, cAMP.J Biol Chem. 2008 Jul 11;283(28):19657-64. doi: 10.1074/jbc.M800595200. Epub 2008 May 13. J Biol Chem. 2008. PMID: 18477562
-
Changes in purine specificity in tandem GAF chimeras from cyanobacterial cyaB1 adenylate cyclase and rat phosphodiesterase 2.FEBS J. 2007 Mar;274(6):1514-23. doi: 10.1111/j.1742-4658.2007.05700.x. FEBS J. 2007. PMID: 17302738
-
Structural and biochemical aspects of tandem GAF domains.Handb Exp Pharmacol. 2009;(191):93-109. doi: 10.1007/978-3-540-68964-5_6. Handb Exp Pharmacol. 2009. PMID: 19089327 Review.
-
Sodium regulation of GAF domain function.Biochem Soc Trans. 2007 Nov;35(Pt 5):1032-4. doi: 10.1042/BST0351032. Biochem Soc Trans. 2007. PMID: 17956270 Review.
Cited by
-
The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change.J Biol Chem. 2008 Sep 19;283(38):25913-9. doi: 10.1074/jbc.M802891200. Epub 2008 Jul 9. J Biol Chem. 2008. PMID: 18614542 Free PMC article.
-
Phytochrome structure and signaling mechanisms.Annu Rev Plant Biol. 2006;57:837-58. doi: 10.1146/annurev.arplant.56.032604.144208. Annu Rev Plant Biol. 2006. PMID: 16669784 Free PMC article. Review.
-
Inactivation of cyclic Di-GMP binding protein TDE0214 affects the motility, biofilm formation, and virulence of Treponema denticola.J Bacteriol. 2013 Sep;195(17):3897-905. doi: 10.1128/JB.00610-13. Epub 2013 Jun 21. J Bacteriol. 2013. PMID: 23794624 Free PMC article.
-
Nitrate- and Nitrite-Sensing Histidine Kinases: Function, Structure, and Natural Diversity.Int J Mol Sci. 2021 May 31;22(11):5933. doi: 10.3390/ijms22115933. Int J Mol Sci. 2021. PMID: 34072989 Free PMC article. Review.
-
Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.J Biol Chem. 2010 Dec 3;285(49):38149-56. doi: 10.1074/jbc.M110.141614. Epub 2010 Sep 21. J Biol Chem. 2010. PMID: 20861010 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

