Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products
- PMID: 15708995
- PMCID: PMC548438
- DOI: 10.1128/JVI.79.5.2768-2779.2005
Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products
Abstract
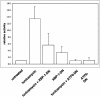
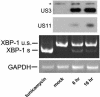
The human cytomegalovirus (HCMV) glycoprotein US11 diverts class I major histocompatibility complex (MHC) heavy chains (HC) from the endoplasmic reticulum (ER) to the cytosol, where HC are subjected to proteasome-mediated degradation. In mouse embryonic fibroblasts that are deficient for X-box binding protein 1 (XBP-1), a key transcription factor in the unfolded protein response (UPR) pathway, we show that degradation of endogenous mouse HC is impaired. Moreover, the rate of US11-mediated degradation of ectopically expressed HLA-A2 is reduced when XBP-1 is absent. In the human astrocytoma cell line U373, turning on expression of US11, but not US2, is sufficient to induce a UPR, as manifested by upregulation of the ER chaperone Bip and by splicing of XBP-1 mRNA. In the presence of dominant-negative versions of XBP-1 and activating transcription factor 6, the kinetics of class I MHC HC degradation were delayed when expression of US11 was turned on. The magnitude of these effects, while reproducible, was modest. Conversely, in cells that stably express high levels of US11, the degradation of HC is not affected by the presence of the dominant negative effectors of the UPR. An infection of human foreskin fibroblasts with human cytomegalovirus induced XBP-1 splicing in a manner that coincides with US11 expression. We conclude that the contribution of the UPR is more pronounced on HC degradation shortly after induction of US11 expression and that US11 is sufficient to induce such a response.
Figures




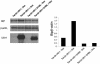

Similar articles
-
Human cytomegalovirus-encoded US2 and US11 target unassembled MHC class I heavy chains for degradation.Mol Immunol. 2006 Mar;43(8):1258-66. doi: 10.1016/j.molimm.2005.07.005. Epub 2005 Aug 10. Mol Immunol. 2006. PMID: 16098592
-
Membrane-specific, host-derived factors are required for US2- and US11-mediated degradation of major histocompatibility complex class I molecules.J Biol Chem. 2002 Feb 1;277(5):3258-67. doi: 10.1074/jbc.M109765200. Epub 2001 Nov 20. J Biol Chem. 2002. PMID: 11717308
-
The role of BiP in endoplasmic reticulum-associated degradation of major histocompatibility complex class I heavy chain induced by cytomegalovirus proteins.J Biol Chem. 2006 Jul 28;281(30):20910-20919. doi: 10.1074/jbc.M602989200. Epub 2006 May 25. J Biol Chem. 2006. PMID: 16731524
-
The HCMV gene products US2 and US11 target MHC class I molecules for degradation in the cytosol.Curr Top Microbiol Immunol. 2002;269:37-55. doi: 10.1007/978-3-642-59421-2_3. Curr Top Microbiol Immunol. 2002. PMID: 12224515 Review.
-
Inhibition of the MHC class II antigen presentation pathway by human cytomegalovirus.Curr Top Microbiol Immunol. 2002;269:101-15. doi: 10.1007/978-3-642-59421-2_7. Curr Top Microbiol Immunol. 2002. PMID: 12224504 Review.
Cited by
-
Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly.J Virol. 2008 Jan;82(1):31-9. doi: 10.1128/JVI.01881-07. Epub 2007 Oct 17. J Virol. 2008. PMID: 17942541 Free PMC article.
-
Human cytomegalovirus induces the endoplasmic reticulum chaperone BiP through increased transcription and activation of translation by using the BiP internal ribosome entry site.J Virol. 2010 Nov;84(21):11479-86. doi: 10.1128/JVI.01330-10. Epub 2010 Aug 25. J Virol. 2010. PMID: 20739513 Free PMC article.
-
Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection.Mol Cell Proteomics. 2011 Oct;10(10):M111.009936. doi: 10.1074/mcp.M111.009936. Epub 2011 Jul 8. Mol Cell Proteomics. 2011. PMID: 21742798 Free PMC article.
-
HCMV Displays a Unique Transcriptome of Immunomodulatory Genes in Primary Monocyte-Derived Cell Types.PLoS One. 2016 Oct 19;11(10):e0164843. doi: 10.1371/journal.pone.0164843. eCollection 2016. PLoS One. 2016. PMID: 27760232 Free PMC article.
-
Feeling manipulated: cytomegalovirus immune manipulation.Virol J. 2009 Jan 9;6:4. doi: 10.1186/1743-422X-6-4. Virol J. 2009. PMID: 19134204 Free PMC article.
References
-
- Casagrande, R., P. Stern, M. Diehn, C. Shamu, M. Osario, M. Zuniga, P. O. Brown, and H. Ploegh. 2000. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell. 5:729-735. - PubMed
-
- Ellgaard, L., and A. Helenius. 2001. ER quality control: towards an understanding at the molecular level. Curr. Opin. Cell Biol. 13:431-437. - PubMed
-
- Friedlander, R., E. Jarosch, J. Urban, C. Volkwein, and T. Sommer. 2000. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2:379-384. - PubMed
-
- Furman, M. H., J. Loureiro, H. L. Ploegh, and D. Tortorella. 2003. Ubiquitinylation of the cytosolic domain of a type I membrane protein is not required to initiate its dislocation from the endoplasmic reticulum. J. Biol. Chem. 278:34804-34811. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

