Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls
- PMID: 15709000
- PMCID: PMC548452
- DOI: 10.1128/JVI.79.5.2814-2822.2005
Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls
Abstract
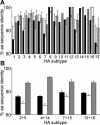
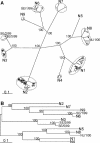
In wild aquatic birds and poultry around the world, influenza A viruses carrying 15 antigenic subtypes of hemagglutinin (HA) and 9 antigenic subtypes of neuraminidase (NA) have been described. Here we describe a previously unidentified antigenic subtype of HA (H16), detected in viruses circulating in black-headed gulls in Sweden. In agreement with established criteria for the definition of antigenic subtypes, hemagglutination inhibition assays and immunodiffusion assays failed to detect specific reactivity between H16 and the previously described subtypes H1 to H15. Genetically, H16 HA was found to be distantly related to H13 HA, a subtype also detected exclusively in shorebirds, and the amino acid composition of the putative receptor-binding site of H13 and H16 HAs was found to be distinct from that in HA subtypes circulating in ducks and geese. The H16 viruses contained NA genes that were similar to those of other Eurasian shorebirds but genetically distinct from N3 genes detected in other birds and geographical locations. The European gull viruses were further distinguishable from other influenza A viruses based on their PB2, NP, and NS genes. Gaining information on the full spectrum of avian influenza A viruses and creating reagents for their detection and identification will remain an important task for influenza surveillance, outbreak control, and animal and public health. We propose that sequence analyses of HA and NA genes of influenza A viruses be used for the rapid identification of existing and novel HA and NA subtypes.
Figures




References
-
- Bokhout, B. A., C. van Gaalen, and P. J. van der Heijden. 1981. A selected water-in-oil emulsion: composition and usefulness as an immunological adjuvant. Vet. Immunol. Immunopathol. 2:491-500.
-
- Chambers, T. M., S. Yamnikova, Y. Kawaoka, D. K. Lvov, and R. G. Webster. 1989. Antigenic and molecular characterization of subtype H13 hemagglutinin of influenza virus. Virology 172:180-188. - PubMed
-
- Cox, N. J., F. Fuller, N. Kaverin, H. D. Klenk, R. A. Lamb, B. W. J. Mahy, J. McCauley, K. Nakamura, P. Palese, and R. Webster. 2000. Orthomyxoviridae, p. 585-597. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
-
- Felsenstein, J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164-166.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

