Quasispecies heterogeneity within the E1/E2 region as a pretreatment variable during pegylated interferon therapy of chronic hepatitis C virus infection
- PMID: 15709027
- PMCID: PMC548442
- DOI: 10.1128/JVI.79.5.3071-3083.2005
Quasispecies heterogeneity within the E1/E2 region as a pretreatment variable during pegylated interferon therapy of chronic hepatitis C virus infection
Abstract
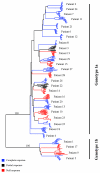
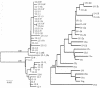
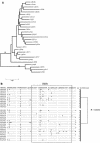
A series of 29 patients undergoing treatment for chronic hepatitis C virus (HCV) genotype 1 infection with pegylated alpha-2a interferon plus ribavirin were studied for patterns of response to antiviral therapy and viral quasispecies evolution. All patients were treatment naive and had chronic inflammation and fibrosis on biopsy. As part of an analysis of pretreatment variables that might affect the outcome of treatment, genetic heterogeneity within the viral E1-E2 glycoprotein region (nucleotides 851 to 2280) was assessed by sequencing 10 to 15 quasispecies clones per patient from serum-derived PCR products. Genetic parameters were examined with respect to response to therapy based on serum viral RNA loads at 12 weeks (early viral response) and at 24 weeks posttreatment (sustained viral response). Nucleotide and amino acid quasispecies complexities of the hypervariable region 1 (HVR-1) were less in the responder group in comparison to the nonresponder group at 12 weeks, and genetic diversity was also less both within and outside of the HVR-1, with the difference being most pronounced for the non-HVR-1 region of E2. However, these genetic parameters did not distinguish responders from nonresponders for sustained viral responses. Follow-up studies of genetic heterogeneity based on the HVR-1 in selected responders and nonresponders while on therapy revealed greater evolutionary drift in the responder subgroup. The pretreatment population sequences for the NS5A interferon sensitivity determinant region were also analyzed for all patients, but no correlations were found between treatment response and any distinct genetic markers. These findings support previous studies indicating a high level of genetic heterogeneity among chronically infected HCV patients. One interpretation of these data is that early viral responses are governed to some extent by viral factors, whereas sustained responses may be more influenced by host factors, in addition to effects of viral complexity and diversity.
Figures
References
-
- Abbate, I., G. Cappiello, O. Lo Iacono, R. Longo, D. Ferraro, G. Antonucci, V. Di Marco, R. Di Stefano, A. Craxi, M. C. Solmone, A. Spano, G. Ippolito, and M. R. Capobianchi. 2003. Heterogeneity of HVR-1 quasispecies is predictive of early but not sustained virological response in genotype 1b-infected patients undergoing combined treatment with PEG- or STD-IFN plus RBV. J. Biol. Regul. Homeost. Agents 17:162-165. - PubMed
-
- Abbate, I., O. Lo Iacono, R. Di Stefano, G. Cappiello, E. Girardi, R. Longo, D. Ferraro, G. Antonucci, V. Di Marco, M. Solmone, A. Craxi, G. Ippolito, and M. R. Capobianchi. 2004. HVR-1 quasispecies modifications occur early and are correlated to initial but not sustained response in HCV-infected patients treated with pegylated- or standard-interferon and ribavirin. J. Hepatol. 40:831-836. - PubMed
-
- Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. - PubMed
-
- Brown, R. S., Jr., and P. J. Gaglio. 2003. Scope of worldwide hepatitis C problem. Liver Transpl. 11:S10-S13. - PubMed
-
- Chayama, K., A. Tsubota, M. Kobayashi, K. Okamoto, M. Hashimoto, Y. Miyano, H. Koike, M. Kobayashi, I. Koida, Y. Arase, S. Saitoh, Y. Suzuki, N. Murashima, K. Ikeda, and H. Kumada. 1997. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology 25:745-749. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

