Interferon regulatory factor 3-independent double-stranded RNA-induced inhibition of hepatitis C virus replicons in human embryonic kidney 293 cells
- PMID: 15709037
- PMCID: PMC548449
- DOI: 10.1128/JVI.79.5.3174-3178.2005
Interferon regulatory factor 3-independent double-stranded RNA-induced inhibition of hepatitis C virus replicons in human embryonic kidney 293 cells
Abstract
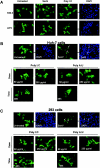
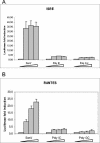
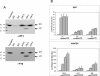
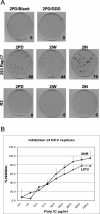
The treatment of human embryonic kidney 293 cells harboring a hepatitis C virus (HCV) subgenomic replicon with the double-stranded RNA (dsRNA) mimic poly(I . C) inhibits HCV RNA replication through an undefined mechanism. Interferon regulatory factor 3 (IRF 3) has been widely postulated to mediate various antiviral responses, and its role in mediating the response to dsRNA in 293 cells was examined. Treating the cells with dsRNA did not induce IRF-3 activation, as measured by nuclear localization or the induction of reporter genes. Moreover, the expression of a dominant negative form of IRF-3 did not affect either colony formation upon transfection of subgenomic replicon RNA or the inhibition of the HCV replicon by dsRNA. Our results suggest that the inhibition of HCV RNA replication by poly(I . C) in 293 cells is independent of IRF-3 activation.
Figures
References
-
- Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732-738. - PubMed
-
- Au, W. C., P. A. Moore, W. Lowther, Y. T. Juang, and P. M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. USA 92:11657-11661. - PMC - PubMed
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Daly, C., and N. C. Reich. 1995. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J. Biol. Chem. 270:23739-23746. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

