Comparative genome analysis of cortactin and HS1: the significance of the F-actin binding repeat domain
- PMID: 15710041
- PMCID: PMC554100
- DOI: 10.1186/1471-2164-6-15
Comparative genome analysis of cortactin and HS1: the significance of the F-actin binding repeat domain
Abstract
Background: In human carcinomas, overexpression of cortactin correlates with poor prognosis. Cortactin is an F-actin-binding protein involved in cytoskeletal rearrangements and cell migration by promoting actin-related protein (Arp)2/3 mediated actin polymerization. It shares a high amino acid sequence and structural similarity to hematopoietic lineage cell-specific protein 1 (HS1) although their functions differ considerable. In this manuscript we describe the genomic organization of these two genes in a variety of species by a combination of cloning and database searches. Based on our analysis, we predict the genesis of the actin-binding repeat domain during evolution.
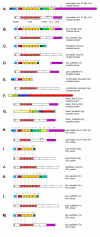
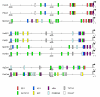
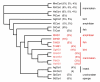
Results: Cortactin homologues exist in sponges, worms, shrimps, insects, urochordates, fishes, amphibians, birds and mammalians, whereas HS1 exists in vertebrates only, suggesting that both genes have been derived from an ancestor cortactin gene by duplication. In agreement with this, comparative genome analysis revealed very similar exon-intron structures and sequence homologies, especially over the regions that encode the characteristic highly conserved F-actin-binding repeat domain. Cortactin splice variants affecting this F-actin-binding domain were identified not only in mammalians, but also in amphibians, fishes and birds. In mammalians, cortactin is ubiquitously expressed except in hematopoietic cells, whereas HS1 is mainly expressed in hematopoietic cells. In accordance with their distinct tissue specificity, the putative promoter region of cortactin is different from HS1.
Conclusions: Comparative analysis of the genomic organization and amino acid sequences of cortactin and HS1 provides inside into their origin and evolution. Our analysis shows that both genes originated from a gene duplication event and subsequently HS1 lost two repeats, whereas cortactin gained one repeat. Our analysis genetically underscores the significance of the F-actin binding domain in cytoskeletal remodeling, which is of importance for the major role of HS1 in apoptosis and for cortactin in cell migration.
Figures

References
-
- Genecard cortactin. 2005. http://bioinfo.weizmann.ac.il/cards-bin/carddisp?CTTN&search=EMS1&suff=txt
-
- Zhan X, Hu X, Hampton B, Burgess WH, Friesel R, Maciag T. Murine cortactin is phosphorylated in response to fibroblast growth factor-1 on tyrosine residues late in the G1 phase of the BALB/c 3T3 cell cycle. J Biol Chem. 1993;268:24427–24431. - PubMed
-
- Schuuring E, Verhoeven E, Mooi WJ, Michalides RJ. Identification and cloning of two overexpressed genes, U21B31/PRAD1 and EMS1, within the amplified chromosome 11q13 region in human carcinomas. Oncogene. 1992;7:355–361. - PubMed
-
- Brookes S, Lammie GA, Schuuring E, de Boer C, Michalides R, Dickson C, Peters G. Amplified region of chromosome band 11q13 in breast and squamous cell carcinomas encompasses three CpG islands telomeric of FGF3, including the expressed gene EMS1. Genes Chromosomes Cancer. 1993;6:222–231. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

