Naturally occurring antisense: transcriptional leakage or real overlap?
- PMID: 15710751
- PMCID: PMC551562
- DOI: 10.1101/gr.3308405
Naturally occurring antisense: transcriptional leakage or real overlap?
Abstract
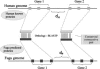
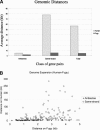
Naturally occurring antisense transcription is associated with the regulation of gene expression through a variety of biological mechanisms. Several recent genome-wide studies reported the identification of potential antisense transcripts for thousands of mammalian genes, many of them resulting from alternatively polyadenylated transcripts or heterogeneous transcription start sites. However, it is not clear whether this transcriptional plasticity is intentional, leading to regulated overlap between the transcripts, or, alternatively, represents a "leakage" of the RNA transcription machinery. To address this question through an evolutionary approach, we compared the genomic organization of genes, with or without antisense, between human, mouse, and the pufferfish Fugu rubripes. Our hypothesis was that if two neighboring genes overlap and have a sense-antisense relationship, we would expect negative selection acting on the evolutionary separation between them. We found that antisense gene pairs are twice as likely to preserve their genomic organization throughout vertebrates' evolution compared to nonantisense pairs, implying an overlap existence in the ancestral genome. In addition, we show that increasing the genomic distance between pairs of genes having a sense-antisense relationship is selected against. These findings indicate that, at least in part, the abundance of antisense transcripts observed in expressed data represents real overlap rather than transcriptional leakage. Moreover, our results imply that natural antisense transcription has considerably affected vertebrate genome evolution.
Figures

References
-
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. - PubMed
-
- Boi, S., Solda G., and Tenchini, M.L. 2004. Shedding light on the dark side of the genome: Overlapping genes in higher eukaryotes. Curr. Genomics 5: 509-524.
-
- Brantl, S. 2002. Antisense-RNA regulation and RNA interference. Biochim. Biophys. Acta 1575: 15-25. - PubMed
-
- Connelly, S. and Manley, J.L. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & Dev. 2: 440-452. - PubMed
WEB SITE REFERENCES
-
- http://www.ncbi.nlm.nih.gov/dbEST/; Expressed Sequence Tags database Web site.
-
- http://www.ncbi.nlm.nih.gov/genome/guide/human/; Human Genome Resources Web site.
-
- http://www.ncbi.nlm.nih.gov/genome/guide/mouse/; Mouse Genome Resources Web site.
-
- http://www.ncbi.nlm.nih.gov/RefSeq/; NCBI Reference Sequences home page.
-
- http://www.ncbi.nlm.nih.gov/HomoloGene/; NCBI Orthologs database Web site.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
