RNA polymerase I transcription factors in active yeast rRNA gene promoters enhance UV damage formation and inhibit repair
- PMID: 15713619
- PMCID: PMC549387
- DOI: 10.1128/MCB.25.5.1586-1595.2005
RNA polymerase I transcription factors in active yeast rRNA gene promoters enhance UV damage formation and inhibit repair
Abstract
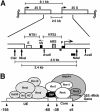
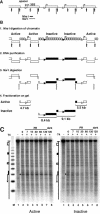
UV photofootprinting and repair of pyrimidine dimers by photolyase was used to investigate chromatin structure, protein-DNA interactions, and DNA repair in the spacer and promoter of Saccharomyces cerevisiae rRNA genes. Saccharomyces cerevisiae contains about 150 copies of rRNA genes separated by nontranscribed spacers. Under exponential growth conditions about half of the genes are transcribed by RNA polymerase I (RNAP-I). Initiation of transcription requires the assembly of the upstream activating factor (UAF), the core factor (CF), TATA binding protein, and RNAP-I with Rrn3p on the upstream element and core promoter. We show that UV irradiation of wild-type cells and transcription factor mutants generates photofootprints in the promoter elements. The core footprint depends on UAF, while the UAF footprint was also detected in absence of the CFs. Fractionation of active and inactive promoters showed the core footprint mainly in the active fraction and similar UAF footprints in both fractions. DNA repair by photolyase was strongly inhibited in active promoters but efficient in inactive promoters. The data suggest that UAF is present in vivo in active and inactive promoters and that recruitment of CF and RNAP-I to active promoters generates a stable complex which inhibits repair.
Figures




References
-
- Becker, M. M., and J. C. Wang. 1984. Use of light for footprinting DNA in vivo. Nature 309:682-687. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
