Displacement of SATB1-bound histone deacetylase 1 corepressor by the human immunodeficiency virus type 1 transactivator induces expression of interleukin-2 and its receptor in T cells
- PMID: 15713622
- PMCID: PMC549366
- DOI: 10.1128/MCB.25.5.1620-1633.2005
Displacement of SATB1-bound histone deacetylase 1 corepressor by the human immunodeficiency virus type 1 transactivator induces expression of interleukin-2 and its receptor in T cells
Erratum in
-
Correction for Kumar et al., "Displacement of SATB1-Bound Histone Deacetylase 1 Corepressor by the Human Immunodeficiency Virus Type 1 Transactivator Induces Expression of Interleukin-2 and Its Receptor in T Cells".Mol Cell Biol. 2022 Apr 21;42(4):e0004022. doi: 10.1128/mcb.00040-22. Epub 2022 Mar 17. Mol Cell Biol. 2022. PMID: 35297674 Free PMC article. No abstract available.
Abstract
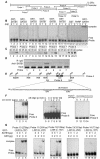
One hallmark of human immunodeficiency virus type 1 (HIV-1) infection is the dysregulation of cytokine gene expression in T cells. Transfection of T cells with human T-cell leukemia type 1 or 2 transactivator results in the induction of the T-cell-restricted cytokine interleukin-2 (IL-2) and its receptor (IL-2Ralpha). However, no T-cell-specific factor(s) has been directly linked with the regulation of IL-2 and IL-2Ralpha transcription by influencing the promoter activity. Thymocytes from SATB1 (special AT-rich sequence binding protein 1) knockout mice have been shown to ectopically express IL-2Ralpha, suggesting involvement of SATB1 in its negative regulation. Here we show that SATB1, a T-cell-specific global gene regulator, binds to the promoters of human IL-2 and IL-2Ralpha and recruits histone deacetylase 1 (HDAC1) in vivo. SATB1 also interacts with Tat in HIV-1-infected T cells. The functional interaction between HIV-1 Tat and SATB1 requires its PDZ-like domain, and the binding of the HDAC1 corepressor occurs through the same. Furthermore, Tat competitively displaces HDAC1 that is bound to SATB1, leading to increased acetylation of the promoters in vivo. Transduction with SATB1 interaction-deficient soluble Tat (Tat 40-72) and reporter assays using a transactivation-negative mutant (C22G) of Tat unequivocally demonstrated that the displacement of HDAC1 itself is sufficient for derepression of these promoters in vivo. These results suggest a novel mechanism by which HIV-1 Tat might overcome SATB1-mediated repression in T cells.
Figures

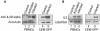






References
-
- Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
