The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis
- PMID: 15713625
- PMCID: PMC549362
- DOI: 10.1128/MCB.25.5.1655-1668.2005
The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis
Abstract
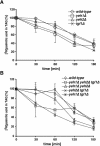
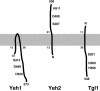
Sterol homeostasis in eukaryotic cells relies on the reciprocal interconversion of free sterols and steryl esters. The formation of steryl esters is well characterized, but the mechanisms that control steryl ester mobilization upon cellular demand are less well understood. We have identified a family of three lipases of Saccharomyces cerevisiae that are required for efficient steryl ester mobilization. These lipases, encoded by YLL012/YEH1, YLR020/YEH2, and TGL1, are paralogues of the mammalian acid lipase family, which is composed of the lysosomal acid lipase, the gastric lipase, and four novel as yet uncharacterized human open reading frames. Lipase triple-mutant yeast cells are completely blocked in steryl ester hydrolysis but do not affect the mobilization of triacylglycerols, indicating that the three lipases are required for steryl ester mobilization in vivo. Lipase single mutants mobilize steryl esters to various degrees, indicating partial functional redundancy of the three gene products. Lipase double-mutant cells in which the third lipase is expressed from the inducible GAL1 promoter have greatly reduced steady-state levels of steryl esters, indicating that overexpression of any of the three lipases is sufficient for steryl ester mobilization in vivo. The three yeast enzymes constitute a novel class of membrane-anchored lipases that differ in topology and subcellular localization.
Figures







Similar articles
-
Yeh1 constitutes the major steryl ester hydrolase under heme-deficient conditions in Saccharomyces cerevisiae.Eukaryot Cell. 2006 Jul;5(7):1018-25. doi: 10.1128/EC.00002-06. Eukaryot Cell. 2006. PMID: 16835446 Free PMC article.
-
The lipid droplet enzyme Tgl1p hydrolyzes both steryl esters and triglycerides in the yeast, Saccharomyces cerevisiae.Biochim Biophys Acta. 2005 Jun 15;1735(1):50-8. doi: 10.1016/j.bbalip.2005.04.005. Biochim Biophys Acta. 2005. PMID: 15922657
-
YEH2/YLR020c encodes a novel steryl ester hydrolase of the yeast Saccharomyces cerevisiae.J Biol Chem. 2005 Apr 8;280(14):13321-8. doi: 10.1074/jbc.M409914200. Epub 2005 Jan 4. J Biol Chem. 2005. PMID: 15632184
-
Formation and mobilization of neutral lipids in the yeast Saccharomyces cerevisiae.Biochem Soc Trans. 2005 Nov;33(Pt 5):1174-7. doi: 10.1042/BST20051174. Biochem Soc Trans. 2005. PMID: 16246075 Review.
-
Synthesis and turnover of non-polar lipids in yeast.Prog Lipid Res. 2008 May;47(3):157-71. doi: 10.1016/j.plipres.2008.01.001. Epub 2008 Jan 18. Prog Lipid Res. 2008. PMID: 18258205 Review.
Cited by
-
A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism.PLoS One. 2010 Oct 28;5(10):e13692. doi: 10.1371/journal.pone.0013692. PLoS One. 2010. PMID: 21060891 Free PMC article.
-
Screening for hydrolytic enzymes reveals Ayr1p as a novel triacylglycerol lipase in Saccharomyces cerevisiae.J Biol Chem. 2013 Dec 13;288(50):36061-72. doi: 10.1074/jbc.M113.509927. Epub 2013 Nov 1. J Biol Chem. 2013. PMID: 24187129 Free PMC article.
-
Lipid droplet dynamics in budding yeast.Cell Mol Life Sci. 2015 Jul;72(14):2677-95. doi: 10.1007/s00018-015-1903-5. Epub 2015 Apr 18. Cell Mol Life Sci. 2015. PMID: 25894691 Free PMC article. Review.
-
Mechanisms by which PE21, an extract from the white willow Salix alba, delays chronological aging in budding yeast.Oncotarget. 2019 Oct 8;10(56):5780-5816. doi: 10.18632/oncotarget.27209. eCollection 2019 Oct 8. Oncotarget. 2019. PMID: 31645900 Free PMC article.
-
Lipid droplet autophagy during energy mobilization, lipid homeostasis and protein quality control.Front Biosci (Landmark Ed). 2018 Mar 1;23(8):1552-1563. doi: 10.2741/4660. Front Biosci (Landmark Ed). 2018. PMID: 29293450 Free PMC article. Review.
References
-
- Abraham, P. R., A. Mulder, J. Van 't Riet, R. J. Planta, and H. A. Raue. 1992. Molecular cloning and physical analysis of an 8.2 kb segment of chromosome XI of Saccharomyces cerevisiae reveals five tightly linked genes. Yeast 8:227-238. - PubMed
-
- Ameis, D., M. Merkel, C. Eckerskorn, and H. Greten. 1994. Purification, characterization and molecular cloning of human hepatic lysosomal acid lipase. Eur. J. Biochem. 219:905-914. - PubMed
-
- Anderson, R. A., and G. N. Sando. 1991. Cloning and expression of cDNA encoding human lysosomal acid lipase/cholesteryl ester hydrolase. Similarities to gastric and lingual lipases. J. Biol. Chem. 266:22479-22484. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
