Modulation of histone deposition by the karyopherin kap114
- PMID: 15713633
- PMCID: PMC549354
- DOI: 10.1128/MCB.25.5.1764-1778.2005
Modulation of histone deposition by the karyopherin kap114
Abstract
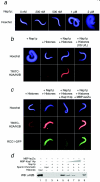
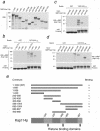
The nuclear import of histones is a prerequisite for the downstream deposition of histones to form chromatin. However, the coordinate regulation of these processes remains poorly understood. Here we demonstrate that Kap114p, the primary karyopherin/importin responsible for the nuclear import of histones H2A and H2B, modulates the deposition of histones H2A and H2B by the histone chaperone Nap1p. We show that a complex comprising Kap114p, histones H2A and H2B, and Nap1p is present in the nucleus and that the presence of this complex is specifically promoted by Nap1p. This places Kap114p in a position to modulate Nap1p function, and we demonstrate by the use of two different assay systems that Kap114p inhibits Nap1p-mediated chromatin assembly. The inhibition of H2A and H2B deposition by Kap114p results in the concomitant inhibition of RCC1 loading onto chromatin. Biochemical evidence suggests that the mechanism by which Kap114p modulates histone deposition primarily involves direct histone binding, while the interaction between Kap114p and Nap1p plays a secondary role. Furthermore, we found that the inhibition of histone deposition by Kap114p is partially reversed by RanGTP. Our results indicate a novel mechanism by which cells can regulate histone deposition and establish a coordinate link between histone nuclear import and chromatin assembly.
Figures









References
-
- Aitchison, J. D., G. Blobel, and M. P. Rout. 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274:624-627. - PubMed
-
- Belotserkovskaya, R., and D. Reinberg. 2004. Facts about FACT and transcript elongation through chromatin. Curr. Opin. Genet. Dev. 14:139-146. - PubMed
-
- Bilbao-Cortes, D., M. Hetzer, G. Langst, P. B. Becker, and I. W. Mattaj. 2002. Ran binds to chromatin by two distinct mechanisms. Curr. Biol. 12:1151-1156. - PubMed
-
- Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427-439. - PubMed
-
- Chang, L., S. S. Loranger, C. Mizzen, S. G. Ernst, C. D. Allis, and A. T. Annunziato. 1997. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry 36:469-480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
