Genetic deletion of the repressor of estrogen receptor activity (REA) enhances the response to estrogen in target tissues in vivo
- PMID: 15713652
- PMCID: PMC549370
- DOI: 10.1128/MCB.25.5.1989-1999.2005
Genetic deletion of the repressor of estrogen receptor activity (REA) enhances the response to estrogen in target tissues in vivo
Abstract
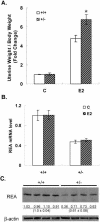
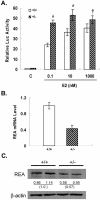
We previously identified a coregulator, repressor of estrogen receptor activity (REA), that directly interacts with estrogen receptor (ER) and represses ER transcriptional activity. Decreasing the intracellular level of REA by using small interfering RNA knockdown or antisense RNA approaches in cells in culture resulted in a significant increase in the level of up-regulation of estrogen-stimulated genes. To elucidate the functional activities of REA in vivo, we have used targeted disruption to delete the REA gene in mice. The targeting vector eliminated, by homologous recombination, the REA exon sequences encoding amino acids 12 to 201, which are required for REA repressive activity and for interaction with ER. The viability of heterozygous animals was similar to that of the wild type, whereas homozygous animals did not develop, suggesting a crucial role for REA in early development. Female, but not male, heterozygous animals had an increased body weight relative to age-matched wild-type animals beginning after puberty. REA mRNA and protein levels in uteri of heterozygous animals were half that of the wild type, and studies with heterozygous animals revealed a greater uterine weight gain and epithelial hyperproliferation in response to estradiol (E2) and a substantially greater stimulation by E2 of a number of estrogen up-regulated genes in the uterus. Even more dramatic in REA heterozygous animals was the loss of down regulation by E2 of genes in the uterus that are normally repressed by estrogen in wild-type animals. Mouse embryo fibroblasts derived from heterozygous embryos also displayed a greater transcriptional response to E2. These studies demonstrate that REA is a significant modulator of estrogen responsiveness in vivo: it normally restrains estrogen actions, moderating ER stimulation and enhancing ER repression of E2-regulated genes.
Figures





Similar articles
-
Analysis of transcription and estrogen insensitivity in the female mouse after targeted disruption of the estrogen receptor gene.Mol Endocrinol. 1995 Nov;9(11):1441-54. doi: 10.1210/mend.9.11.8584021. Mol Endocrinol. 1995. PMID: 8584021
-
Methoxychlor stimulates estrogen-responsive messenger ribonucleic acids in mouse uterus through a non-estrogen receptor (non-ER) alpha and non-ER beta mechanism.Endocrinology. 1999 Aug;140(8):3526-33. doi: 10.1210/endo.140.8.6877. Endocrinology. 1999. PMID: 10433208
-
The coregulator, repressor of estrogen receptor activity (REA), is a crucial regulator of the timing and magnitude of uterine decidualization.Endocrinology. 2013 Mar;154(3):1349-60. doi: 10.1210/en.2012-2026. Epub 2013 Feb 7. Endocrinology. 2013. PMID: 23392257 Free PMC article.
-
Estrogen receptor gene disruption: molecular characterization and experimental and clinical phenotypes.Recent Prog Horm Res. 1996;51:159-86; discussion 186-8. Recent Prog Horm Res. 1996. PMID: 8701078 Review.
-
Modulation of estrogen receptor activity by selective coregulators.J Steroid Biochem Mol Biol. 2003 Jun;85(2-5):117-22. doi: 10.1016/s0960-0760(03)00207-3. J Steroid Biochem Mol Biol. 2003. PMID: 12943695 Review.
Cited by
-
Uterine development and fertility are dependent on gene dosage of the nuclear receptor coregulator REA.Endocrinology. 2012 Aug;153(8):3982-94. doi: 10.1210/en.2012-1044. Epub 2012 May 14. Endocrinology. 2012. PMID: 22585830 Free PMC article.
-
Estradiol inhibits Th17 cell differentiation through inhibition of RORγT transcription by recruiting the ERα/REA complex to estrogen response elements of the RORγT promoter.J Immunol. 2015 Apr 15;194(8):4019-28. doi: 10.4049/jimmunol.1400806. Epub 2015 Mar 13. J Immunol. 2015. PMID: 25769926 Free PMC article.
-
The Prohibitins: emerging roles in diverse functions.J Cell Mol Med. 2006 Apr-Jun;10(2):353-63. doi: 10.1111/j.1582-4934.2006.tb00404.x. J Cell Mol Med. 2006. PMID: 16796804 Free PMC article. Review.
-
Corepressors of agonist-bound nuclear receptors.Toxicol Appl Pharmacol. 2007 Sep 15;223(3):288-98. doi: 10.1016/j.taap.2007.05.019. Epub 2007 Jun 14. Toxicol Appl Pharmacol. 2007. PMID: 17628626 Free PMC article. Review.
-
Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria.Genes Dev. 2008 Feb 15;22(4):476-88. doi: 10.1101/gad.460708. Genes Dev. 2008. PMID: 18281461 Free PMC article.
References
-
- Ansari-Lari, M. A., J. C. Oeltjen, S. Schwartz, Z. Zhang, D. M. Muzny, J. Lu, J. H. Gorrell, A. C. Chinault, J. W. Belmont, W. Miller, and R. A. Gibbs. 1998. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 8:29-40. - PubMed
-
- Ansari-Lari, M. A., Y. Shen, D. M. Muzny, W. Lee, and R. A. Gibbs. 1997. Large-scale sequencing in human chromosome 12p13: experimental and computational gene structure determination. Genome Res. 7:268-280. - PubMed
-
- Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. - PubMed
-
- Burakov, D., L. A. Crofts, C. P. Chang, and L. P. Freedman. 2002. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J. Biol. Chem. 277:14359-14362. - PubMed
-
- Delage-Mourroux, R., P. G. Martini, I. Choi, D. M. Kraichely, J. Hoeksema, and B. S. Katzenellenbogen. 2000. Analysis of estrogen receptor interaction with a repressor of estrogen receptor activity (REA) and the regulation of estrogen receptor transcriptional activity by REA. J. Biol. Chem. 275:35848-35856. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
