Evolutionary significance of gene expression divergence
- PMID: 15716085
- PMCID: PMC1859841
- DOI: 10.1016/j.gene.2004.11.034
Evolutionary significance of gene expression divergence
Abstract
Recent large-scale studies of evolutionary changes in gene expression among mammalian species have led to the proposal that gene expression divergence may be neutral with respect to organismic fitness. Here, we employ a comparative analysis of mammalian gene sequence divergence and gene expression divergence to test the hypothesis that the evolution of gene expression is predominantly neutral. Two models of neutral gene expression evolution are considered: 1-purely neutral evolution (i.e., no selective constraint) of gene expression levels and patterns and 2-neutral evolution accompanied by selective constraint. With respect to purely neutral evolution, levels of change in gene expression between human-mouse orthologs are correlated with levels of gene sequence divergence that are determined largely by purifying selection. In contrast, evolutionary changes of tissue-specific gene expression profiles do not show such a correlation with sequence divergence. However, divergence of both gene expression levels and profiles are significantly lower for orthologous human-mouse gene pairs than for pairs of randomly chosen human and mouse genes. These data clearly point to the action of selective constraint on gene expression divergence and are inconsistent with the purely neutral model; however, there is likely to be a neutral component in evolution of gene expression, particularly, in tissues where the expression of a given gene is low and functionally irrelevant. The model of neutral evolution with selective constraint predicts a regular, clock-like accumulation of gene expression divergence. However, relative rate tests of the divergence among human-mouse-rat orthologous gene sets reveal clock-like evolution for gene sequence divergence, and to a lesser extent for gene expression level divergence, but not for the divergence of tissue-specific gene expression profiles. Taken together, these results indicate that gene expression divergence is subject to the effects of purifying selective constraint and suggest that it might also be substantially influenced by positive Darwinian selection.
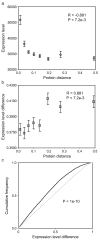
Figures



References
-
- Adams MD, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
-
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. - PubMed
-
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–138. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

