Structural and spectroscopic properties of a reaction center complex from the chlorosome-lacking filamentous anoxygenic phototrophic bacterium Roseiflexus castenholzii
- PMID: 15716441
- PMCID: PMC1063993
- DOI: 10.1128/JB.187.5.1702-1709.2005
Structural and spectroscopic properties of a reaction center complex from the chlorosome-lacking filamentous anoxygenic phototrophic bacterium Roseiflexus castenholzii
Abstract
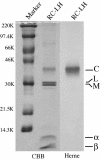
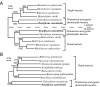
The photochemical reaction center (RC) complex of Roseiflexus castenholzii, which belongs to the filamentous anoxygenic phototrophic bacteria (green filamentous bacteria) but lacks chlorosomes, was isolated and characterized. The genes coding for the subunits of the RC and the light-harvesting proteins were also cloned and sequenced. The RC complex was composed of L, M, and cytochrome subunits. The cytochrome subunit showed a molecular mass of approximately 35 kDa, contained hemes c, and functioned as the electron donor to the photo-oxidized special pair of bacteriochlorophylls in the RC. The RC complex appeared to contain three molecules of bacteriochlorophyll and three molecules of bacteriopheophytin, as in the RC preparation from Chloroflexus aurantiacus. Phylogenetic trees based on the deduced amino acid sequences of the RC subunits suggested that R. castenholzii had diverged from C. aurantiacus very early after the divergence of filamentous anoxygenic phototrophic bacteria from purple bacteria. Although R. castenholzii is phylogenetically related to C. aurantiacus, the arrangement of its puf genes, which code for the light-harvesting proteins and the RC subunits, was different from that in C. aurantiacus and similar to that in purple bacteria. The genes are found in the order pufB, -A, -L, -M, and -C, with the pufL and pufM genes forming one continuous open reading frame. Since the photosynthetic apparatus and genes of R. castenholzii have intermediate characteristics between those of purple bacteria and C. aurantiacus, it is likely that they retain many features of the common ancestor of purple bacteria and filamentous anoxygenic phototrophic bacteria.
Figures


Similar articles
-
Characterization of a blue-copper protein, auracyanin, of the filamentous anoxygenic phototrophic bacterium Roseiflexus castenholzii.Arch Biochem Biophys. 2009 Oct 1;490(1):57-62. doi: 10.1016/j.abb.2009.08.003. Epub 2009 Aug 14. Arch Biochem Biophys. 2009. PMID: 19683508
-
Kinetics and energetics of electron transfer in reaction centers of the photosynthetic bacterium Roseiflexus castenholzii.Biochim Biophys Acta. 2011 Mar;1807(3):262-9. doi: 10.1016/j.bbabio.2010.11.011. Epub 2010 Nov 29. Biochim Biophys Acta. 2011. PMID: 21126505
-
Excitation energy transfer and trapping dynamics in the core complex of the filamentous photosynthetic bacterium Roseiflexus castenholzii.Photosynth Res. 2012 Mar;111(1-2):149-56. doi: 10.1007/s11120-011-9669-6. Epub 2011 Jul 27. Photosynth Res. 2012. PMID: 21792612
-
Seeing green bacteria in a new light: genomics-enabled studies of the photosynthetic apparatus in green sulfur bacteria and filamentous anoxygenic phototrophic bacteria.Arch Microbiol. 2004 Oct;182(4):265-76. doi: 10.1007/s00203-004-0718-9. Epub 2004 Sep 1. Arch Microbiol. 2004. PMID: 15340781 Review.
-
What We Are Learning from the Diverse Structures of the Homodimeric Type I Reaction Center-Photosystems of Anoxygenic Phototropic Bacteria.Biomolecules. 2024 Mar 6;14(3):311. doi: 10.3390/biom14030311. Biomolecules. 2024. PMID: 38540731 Free PMC article. Review.
Cited by
-
A Novel Microbialite-Associated Phototrophic Chloroflexi Lineage Exhibiting a Quasi-Clonal Pattern along Depth.Genome Biol Evol. 2020 Jul 1;12(7):1207-1216. doi: 10.1093/gbe/evaa122. Genome Biol Evol. 2020. PMID: 32544224 Free PMC article.
-
Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote.Nat Commun. 2018 Apr 19;9(1):1568. doi: 10.1038/s41467-018-03881-x. Nat Commun. 2018. PMID: 29674684 Free PMC article.
-
Pigment-modified reaction centers of Chloroflexus aurantiacus: chemical exchange of bacteriopheophytins with plant-type pheophytins.Photosynth Res. 2021 Sep;149(3):313-328. doi: 10.1007/s11120-021-00855-x. Epub 2021 Jun 17. Photosynth Res. 2021. PMID: 34138452
-
Structural basis underlying the electron transfer features of a blue copper protein auracyanin from the photosynthetic bacterium Roseiflexus castenholzii.Photosynth Res. 2020 Mar;143(3):301-314. doi: 10.1007/s11120-020-00709-y. Epub 2020 Jan 13. Photosynth Res. 2020. PMID: 31933173
-
Evolution of Phototrophy in the Chloroflexi Phylum Driven by Horizontal Gene Transfer.Front Microbiol. 2018 Feb 19;9:260. doi: 10.3389/fmicb.2018.00260. eCollection 2018. Front Microbiol. 2018. PMID: 29515543 Free PMC article.
References
-
- Blankenship, R. E., R. Feick, B. D. Bruce, C. Kirmaier, D. Holten, and R. C. Fuller. 1983. Primary photochemistry in the facultative green photosynthetic bacterium Chloroflexus aurantiacus. J. Cell Biochem. 22:251-261. - PubMed
-
- Blankenship, R. E. 1992. Origin and evolution of photosynthesis. Photosynth. Res. 33:91-111. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

