Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes
- PMID: 15718296
- PMCID: PMC549392
- DOI: 10.1093/nar/gki201
Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes
Abstract
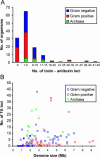
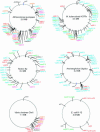
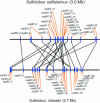
Prokaryotic chromosomes code for toxin-antitoxin (TA) loci, often in multiple copies. In E.coli, experimental evidence indicates that TA loci are stress-response elements that help cells survive unfavorable growth conditions. The first gene in a TA operon codes for an antitoxin that combines with and neutralizes a regulatory 'toxin', encoded by the second gene. RelE and MazF toxins are regulators of translation that cleave mRNA and function, in interplay with tmRNA, in quality control of gene expression. Here, we present the results from an exhaustive search for TA loci in 126 completely sequenced prokaryotic genomes (16 archaea and 110 bacteria). We identified 671 TA loci belonging to the seven known TA gene families. Surprisingly, obligate intracellular organisms were devoid of TA loci, whereas free-living slowly growing prokaryotes had particularly many (38 in Mycobacterium tuberculosis and 43 in Nitrosomonas europaea). In many cases, TA loci were clustered and closely linked to mobile genetic elements. In the most extreme of these cases, all 13 TA loci of Vibrio cholerae were bona fide integron elements located in the V.cholerae mega-integron. These observations strongly suggest that TA loci are mobile cassettes that move frequently within and between chromosomes and also lend support to the hypothesis that TA loci function as stress-response elements beneficial to free-living prokaryotes.
Figures



References
-
- Le Hir H., Nott A., Moore M.J. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 2003;28:215–220. - PubMed
-
- Singh G., Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 2003;28:464–466. - PubMed
-
- Karzai A.W., Roche E.D., Sauer R.T. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nature Struct. Biol. 2000;7:449–455. - PubMed
-
- Keiler K.C., Waller P.R., Sauer R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. - PubMed
-
- Christensen S.K., Pedersen K., Hansen F.G., Gerdes K. Toxin–antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol Biol. 2003;332:809–819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

