Mms22p protects Saccharomyces cerevisiae from DNA damage induced by topoisomerase II
- PMID: 15718301
- PMCID: PMC549411
- DOI: 10.1093/nar/gki246
Mms22p protects Saccharomyces cerevisiae from DNA damage induced by topoisomerase II
Abstract
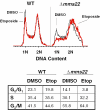
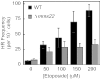
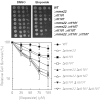
The cleavage reaction of topoisomerase II, which creates double-stranded DNA breaks, plays a central role in both the cure and initiation of cancer. Therefore, it is important to understand the cellular processes that repair topoisomerase II-generated DNA damage. Using a genome-wide approach with Saccharomyces cerevisiae, we found that Deltamre11, Deltaxrs2, Deltarad50, Deltarad51, Deltarad52, Deltarad54, Deltarad55, Deltarad57 and Deltamms22 strains were hypersensitive to etoposide, a drug that specifically increases levels of topoisomerase II-mediated DNA breaks. These results confirm that the single-strand invasion pathway of homologous recombination is the major pathway that repairs topoisomerase II-induced DNA damage in yeast and also indicate an important role for Mms22p. Although Deltamms22 strains are sensitive to several DNA-damaging agents, little is known about the function of Mms22p. Deltamms22 cultures accumulate in G2/M, and display an abnormal cell cycle response to topoisomerase II-mediated DNA damage. MMS22 appears to function outside of the single-strand invasion pathway, but levels of etoposide-induced homologous recombination in Deltamms22 cells are lower than wild-type. MMS22 is epistatic with RTT101 and RTT107, genes that encode its protein binding partners. Finally, consistent with a role in DNA processes, Mms22p localizes to discrete nuclear foci, even in the absence of etoposide or its binding partners.
Figures






References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Jackson A.L., Newcomb T.G., Loeb L.A. Origin of multiple mutations in human cancers. Drug Metab. Rev. 1998;30:285–304. - PubMed
-
- Friedberg E.C. DNA damage and repair. Nature. 2003;421:436–440. - PubMed
-
- Friedberg E.C., Walker G.C., Siede W. DNA Repair and Mutagenesis, 2nd edn. Washington, DC: American Society for Microbiology Press; 1995.
-
- Marnett L.J., Plastaras J.P. Endogenous DNA damage and mutation. Trends Genet. 2001;17:214–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

