Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities
- PMID: 15719014
- PMCID: PMC554134
- DOI: 10.1038/sj.emboj.7600588
Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities
Abstract
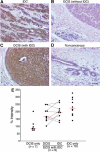
Identification of the molecular machinery employed in cancer invasion, but not in normal adult cells, will greatly contribute to cancer therapeutics. Here we found that an ArfGAP, AMAP1/PAG2, is expressed at high levels in highly invasive breast cancer cells, but at very low levels in noninvasive breast cancer cells and normal mammary epithelial cells. siRNA-mediated silencing of AMAP1 effectively blocked the invasive activities. AMAP1 expression in human breast primary tumors also indicated its potential correlation with malignancy. Paxillin and cortactin have been shown to colocalize at invadopodia and play a pivotal role in breast cancer invasion. We found that AMAP1 is also localized at invadopodia, and acts to bridge paxillin and cortactin. This AMAP1-mediated trimeric protein complex was detected only in invasive cancer cells, and blocking this complex formation effectively inhibited their invasive activities in vitro and metastasis in mice. Our results indicate that AMAP1 is a component involved in invasive activities of different breast cancers, and provide new information regarding the possible therapeutic targets for prevention of breast cancer invasion and metastasis.
Figures







References
-
- Allred DC, Mohsin SK, Fuqua SA (2001) Histological and biological evolution of human premalignant breast disease. Endocr Relat Cancer 8: 47–61 - PubMed
-
- Bowden ET, Barth M, Thomas D, Glazer RI, Mueller SC (1999) An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene 18: 4440–4449 - PubMed
-
- Bowden ET, Coopman PJ, Mueller SC (2001) Invadopodia: unique methods for measurement of extracellular matrix degradation in vitro. Methods Cell Biol 63: 613–627 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

