The Arabidopsis Mei2 homologue AML1 binds AtRaptor1B, the plant homologue of a major regulator of eukaryotic cell growth
- PMID: 15720729
- PMCID: PMC553980
- DOI: 10.1186/1471-2229-5-2
The Arabidopsis Mei2 homologue AML1 binds AtRaptor1B, the plant homologue of a major regulator of eukaryotic cell growth
Abstract
Background: TOR, the target of the antibiotic rapamycin in both yeast and mammalian cells, is a potent cell growth regulator in all eukaryotes. It acts through the phosphorylation of downstream effectors that are recruited to it by the binding partner Raptor. In Arabidopsis, Raptor activity is essential for postembryonic growth. Though comparative studies suggest potential downstream effectors, no Raptor binding partners have been described in plants.
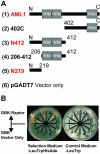
Results: AtRaptor1B, a plant Raptor homologue, binds the AML1 (Arabidopsis Mei2-like 1) protein in a yeast two-hybrid assay. This interaction is mediated by the N-terminal 219 residues of AML1, and marks AML1 as a candidate AtTOR kinase substrate in plants. The AML1 N-terminus additionally carries transcriptional activation domain activity. Plants homozygous for insertion alleles at the AML1 locus, as well as plants homozygous for insertion alleles at all five loci in the AML gene family, bolt earlier than wild-type plants.
Conclusion: AML1 interacts with AtRaptor1B, homologue of a protein that recruits substrates for phosphorylation by the major cell-growth regulator TOR. Identification of AML1 as a putative downstream effector of TOR gives valuable insights into the plant-specific mode of action of this critical growth regulator.
Figures




References
-
- Thomas G, Sabatini D, Hall M. TOR Target of Rapamycin. New York: Springer-Verlag; 2004.
-
- Kim DH, Sarbassov D, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/S1097-2765(03)00114-X. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

