Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data
- PMID: 15720800
- PMCID: PMC1531678
- DOI: 10.1593/neo.04277
Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data
Abstract
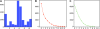
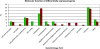
Cancers originating from epithelial cells are the most common malignancies. No common expression profile of solid tumors compared to normal tissues has been described so far. Therefore we were interested if genes differentially expressed in the majority of carcinomas could be identified using bioinformatic methods. Complete data sets were downloaded for carcinomas of the prostate, breast, lung, ovary, colon, pancreas, stomach, bladder, liver, and kidney, and were subjected to an expression analysis using SAM. In each experiment, a gene was scored as differentially expressed if the q value was below 25%. Probe identifiers were unified by comparing the respective probe sequences to the Unigene build 155 using BlastN. To obtain differentially expressed genes within the set of analyzed carcinomas, the number of experiments in which differential expression was observed was counted. Differential expression was assigned to genes if they were differentially expressed in at least eight experiments of tumors from different origin. The identified candidate genes ADRM1, EBNA1BP2, FDPS, FOXM1, H2AFX, HDAC3, IRAK1, and YY1 were subjected to further validation. Using this comparative approach, 100 genes were identified as upregulated and 21 genes as downregulated in the carcinomas.
Figures

References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. - PubMed
-
- Amatschek S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A, Dekan G, Vogl S, Kubista E, Heider KH, et al. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004;64:844–856. - PubMed
-
- Buckhaults P, Zhang Z, Chen YC, Wang TL, St Croix B, Saha S, Bardelli A, Morin PJ, Polyak K, Hruban RH, et al. Identifying tumor origin using a gene expression-based classification map. Cancer Res. 2003;63:4144–4149. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
