Insights into the domains required for dimerization and assembly of human alphaB crystallin
- PMID: 15722445
- PMCID: PMC2279284
- DOI: 10.1110/ps.041152805
Insights into the domains required for dimerization and assembly of human alphaB crystallin
Abstract
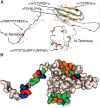
Protein pin array technology was used to identify subunit-subunit interaction sites in the small heat shock protein (sHSP) alphaB crystallin. Subunit-subunit interaction sites were defined as consensus sequences that interacted with both human alphaA crystallin and alphaB crystallin. The human alphaB crystallin protein pin array consisted of contiguous and overlapping peptides, eight amino acids in length, immobilized on pins that were in a 96-well ELISA plate format. The interaction of alphaB crystallin peptides with physiological partner proteins, alphaA crystallin and alphaB crystallin, was detected using antibodies and recorded using spectrophotometric absorbance. Five peptide sequences including 37LFPTSTSLSPFYLRPPSF54 in the N terminus, 75FSVNLDVK82)(beta3), 131LTITSSLS138 (beta8) and 141GVLTVNGP148 (beta9) that form beta strands in the conserved alpha crystallin core domain, and 155PERTIPITREEK166 in the C-terminal extension were identified as subunit-subunit interaction sites in human alphaB crystallin using the novel protein pin array assay. The subunit-subunit interaction sites were mapped to a three-dimensional (3D) homology model of wild-type human alphaB crystallin that was based on the crystal structure of wheat sHSP16.9 and Methanococcus jannaschi sHSP16.5 (Mj sHSP16.5). The subunit-subunit interaction sites identified and mapped onto the homology model were solvent-exposed and had variable secondary structures ranging from beta strands to random coils and short alpha helices. The subunit-subunit interaction sites formed a pattern of hydrophobic patches on the 3D surface of human alphaB crystallin.
Figures


Similar articles
-
Interactive domains for chaperone activity in the small heat shock protein, human alphaB crystallin.Biochemistry. 2005 Nov 15;44(45):14854-69. doi: 10.1021/bi0503910. Biochemistry. 2005. PMID: 16274233
-
AlphaA-crystallin interacting regions in the small heat shock protein, alphaB-crystallin.Biochemistry. 2004 Dec 21;43(50):15785-95. doi: 10.1021/bi048151s. Biochemistry. 2004. PMID: 15595834
-
The function of the beta3 interactive domain in the small heat shock protein and molecular chaperone, human alphaB crystallin.Cell Stress Chaperones. 2006 Summer;11(2):187-97. doi: 10.1379/csc-186.1. Cell Stress Chaperones. 2006. PMID: 16817325 Free PMC article.
-
Functional sequences in human alphaB crystallin.Biochim Biophys Acta. 2016 Jan;1860(1 Pt B):240-5. doi: 10.1016/j.bbagen.2015.08.014. Epub 2015 Sep 2. Biochim Biophys Acta. 2016. PMID: 26341790 Free PMC article. Review.
-
One size does not fit all: the oligomeric states of αB crystallin.FEBS Lett. 2013 Apr 17;587(8):1073-80. doi: 10.1016/j.febslet.2013.01.021. Epub 2013 Jan 20. FEBS Lett. 2013. PMID: 23340341 Free PMC article. Review.
Cited by
-
Structural and functional roles of deamidation of N146 and/or truncation of NH2- or COOH-termini in human αB-crystallin.Mol Vis. 2011;17:2407-20. Epub 2011 Sep 14. Mol Vis. 2011. PMID: 21976952 Free PMC article.
-
Key Role of Phosphorylation in Small Heat Shock Protein Regulation via Oligomeric Disaggregation and Functional Activation.Cells. 2025 Jan 17;14(2):127. doi: 10.3390/cells14020127. Cells. 2025. PMID: 39851555 Free PMC article. Review.
-
Mini-alphaB-crystallin: a functional element of alphaB-crystallin with chaperone-like activity.Biochemistry. 2006 Mar 7;45(9):3069-76. doi: 10.1021/bi0518141. Biochemistry. 2006. PMID: 16503662 Free PMC article.
-
Partially folded aggregation intermediates of human gammaD-, gammaC-, and gammaS-crystallin are recognized and bound by human alphaB-crystallin chaperone.J Mol Biol. 2010 Aug 6;401(1):134-52. doi: 10.1016/j.jmb.2010.05.067. Epub 2010 Jun 1. J Mol Biol. 2010. PMID: 20621668 Free PMC article.
-
Structural and functional impact of the p.R163C mutation in the conserved palindromic motif within the C-terminal domain of human αB-crystallin.PLoS One. 2025 Jul 14;20(7):e0326025. doi: 10.1371/journal.pone.0326025. eCollection 2025. PLoS One. 2025. PMID: 40658662 Free PMC article.
References
-
- Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132 221–241. - PubMed
-
- Alizadeh, A., Clark, J., Seeberger, T., Hess, J., Blankenship, T., and FitzGerald, P.G. 2003. Targeted deletion of the lens fiber cell-specific intermediate filament protein filensin. Invest. Ophthalmol. Vis. Sci. 44 5252–5258. - PubMed
-
- ———. 2004. Characterization of a mutation in the lens-specific CP49 in the 129 strain of mouse. Invest. Ophthalmol. Vis. Sci. 45 884–891. - PubMed
-
- Aquilina, J.A., Benesch, J.L., Ding, L.L., Yaron, O., Horwitz, J., and Robinson, C.V. 2004. Phosphorylation of αB-crystallin alters chaperone function through loss of dimeric substructure. J. Biol. Chem. 279 28675–28680. - PubMed
-
- Bernard, J., Harb, C., Mortier, E., Quemener, A., Meloen, R.H., Vermot-Desroches, C., Wijdeness, J., van Dijken, P., Grotzinger, J., Slootstra, J.W., et al. 2004. Identification of an interleukin-15α receptor-binding site on human interleukin-15. J. Biol. Chem. 279 24313–24322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

