Microsecond timescale backbone conformational dynamics in ubiquitin studied with NMR R1rho relaxation experiments
- PMID: 15722448
- PMCID: PMC2279275
- DOI: 10.1110/ps.041139505
Microsecond timescale backbone conformational dynamics in ubiquitin studied with NMR R1rho relaxation experiments
Abstract
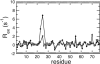
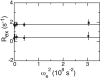
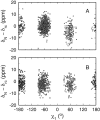
NMR spin relaxation experiments are used to characterize the dynamics of the backbone of ubiquitin. Chemical exchange processes affecting residues Ile 23, Asn 25, Thr 55, and Val 70 are characterized using on- and off-resonance rotating-frame 15N R1rho relaxation experiments to have a kinetic exchange rate constant of 25,000 sec(-1) at 280 K. The exchange process affecting residues 23, 25, and 55 appears to result from disruption of N-cap hydrogen bonds of the alpha-helix and possibly from repacking of the side chain of Ile 23. Chemical exchange processes affecting other residues on the surface of ubiquitin are identified using 1H-15N multiple quantum relaxation experiments. These residues are located near or at the regions known to interact with various enzymes of the ubiquitin-dependent protein degradation pathway.
Figures




References
-
- Abragam, A. 1983. Principles of nuclear magnetism. Oxford University Press, Oxford.
-
- Braun, D., Wider, G., and Wüthrich, K. 1994. Sequence corrected 15N random coil chemical-shifts. J. Am. Chem. Soc. 116 8466–8469.
-
- Burch, T.J. and Haas, A.L. 1994. Site-directed mutagenesis of ubiquitin. Differential roles of arginine in the interaction with ubiquitin-activating enzyme. Biochemistry 33 7300–7308. - PubMed
-
- Carlomagno, T., Maurer, M., Hennig, M., and Griesinger, C. 2000. Ubiquitin backbone motion studied via NHN-C′ Cα dipolar-dipolar and C′-C′ C′/ NHN CSA-dipolar cross-correlated relaxation. J. Am. Chem. Soc. 122 5105–5113.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

