Cell type-specific localization of transcripts encoding nine consecutive enzymes involved in protoberberine alkaloid biosynthesis
- PMID: 15722473
- PMCID: PMC1069708
- DOI: 10.1105/tpc.104.028654
Cell type-specific localization of transcripts encoding nine consecutive enzymes involved in protoberberine alkaloid biosynthesis
Abstract
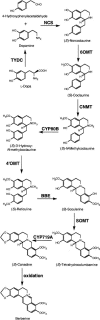
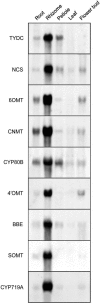
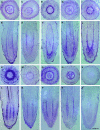
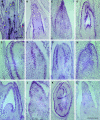
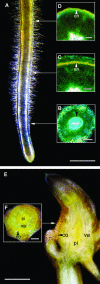
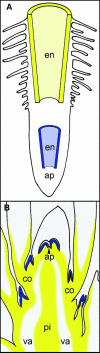
Molecular clones encoding nine consecutive biosynthetic enzymes that catalyze the conversion of l-dopa to the protoberberine alkaloid (S)-canadine were isolated from meadow rue (Thalictrum flavum ssp glaucum). The predicted proteins showed extensive sequence identity with corresponding enzymes involved in the biosynthesis of related benzylisoquinoline alkaloids in other species, such as opium poppy (Papaver somniferum). RNA gel blot hybridization analysis showed that gene transcripts for each enzyme were most abundant in rhizomes but were also detected at lower levels in roots and other organs. In situ RNA hybridization analysis revealed the cell type-specific expression of protoberberine alkaloid biosynthetic genes in roots and rhizomes. In roots, gene transcripts for all nine enzymes were localized to immature endodermis, pericycle, and, in some cases, adjacent cortical cells. In rhizomes, gene transcripts encoding all nine enzymes were restricted to the protoderm of leaf primordia. The localization of biosynthetic gene transcripts was in contrast with the tissue-specific accumulation of protoberberine alkaloids. In roots, protoberberine alkaloids were restricted to mature endodermal cells upon the initiation of secondary growth and were distributed throughout the pith and cortex in rhizomes. Thus, the cell type-specific localization of protoberberine alkaloid biosynthesis and accumulation are temporally and spatially separated in T. flavum roots and rhizomes, respectively. Despite the close phylogeny between corresponding biosynthetic enzymes, distinct and different cell types are involved in the biosynthesis and accumulation of benzylisoquinoline alkaloids in T. flavum and P. somniferum. Our results suggest that the evolution of alkaloid metabolism involves not only the recruitment of new biosynthetic enzymes, but also the migration of established pathways between cell types.
Figures
References
-
- Burlat, V., Oudin, A., Courtois, M., Rideau, M., and St-Pierre, B. (2004). Co-expression of three MEP pathway genes and geraniol10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J. 38, 131–141. - PubMed
-
- Choi, K.B., Morishige, T., Shitan, N., Yazaki, K., and Sato, F. (2002). Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 277, 830–835. - PubMed
-
- Facchini, P.J. (2001). Alkaloid biosynthesis in plants: Biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 29–66. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

