Mutant p53 proteins bind DNA in a DNA structure-selective mode
- PMID: 15722483
- PMCID: PMC549414
- DOI: 10.1093/nar/gki252
Mutant p53 proteins bind DNA in a DNA structure-selective mode
Abstract
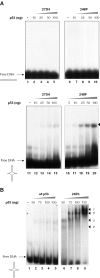
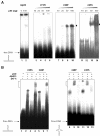
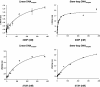
Despite the loss of sequence-specific DNA binding, mutant p53 (mutp53) proteins can induce or repress transcription of mutp53-specific target genes. To date, the molecular basis for transcriptional modulation by mutp53 is not understood, but increasing evidence points to the possibility that specific interactions of mutp53 with DNA play an important role. So far, the lack of a common denominator for mutp53 DNA binding, i.e. the existence of common sequence elements, has hampered further characterization of mutp53 DNA binding. Emanating from our previous discovery that DNA structure is an important determinant of wild-type p53 (wtp53) DNA binding, we analyzed the binding of various mutp53 proteins to oligonucleotides mimicking non-B DNA structures. Using various DNA-binding assays we show that mutp53 proteins bind selectively and with high affinity to non-B DNA. In contrast to sequence-specific and DNA structure-dependent binding of wtp53, mutp53 DNA binding to non-B DNA is solely dependent on the stereo-specific configuration of the DNA, and not on DNA sequence. We propose that DNA structure-selective binding of mutp53 proteins is the basis for the well-documented interaction of mutp53 with MAR elements and for transcriptional activities mediates by mutp53.
Figures




Similar articles
-
APE1/Ref-1 enhances DNA binding activity of mutant p53 in a redox-dependent manner.Oncol Rep. 2014 Feb;31(2):901-9. doi: 10.3892/or.2013.2892. Epub 2013 Dec 2. Oncol Rep. 2014. PMID: 24297337
-
DNA-conformation is an important determinant of sequence-specific DNA binding by tumor suppressor p53.Oncogene. 1997 Aug 14;15(7):857-69. doi: 10.1038/sj.onc.1201412. Oncogene. 1997. PMID: 9266973
-
Restoring wild-type conformation and DNA-binding activity of mutant p53 is insufficient for restoration of transcriptional activity.Biochem Biophys Res Commun. 2006 Dec 15;351(2):499-506. doi: 10.1016/j.bbrc.2006.10.065. Epub 2006 Oct 19. Biochem Biophys Res Commun. 2006. PMID: 17070499
-
Mutant p53: "gain of function" through perturbation of nuclear structure and function?J Cell Biochem Suppl. 2000;Suppl 35:115-22. J Cell Biochem Suppl. 2000. PMID: 11389540 Review.
-
Transcriptional activities of mutant p53: when mutations are more than a loss.J Cell Biochem. 2004 Nov 15;93(5):878-86. doi: 10.1002/jcb.20271. J Cell Biochem. 2004. PMID: 15449312 Review.
Cited by
-
Differential salt-induced dissociation of the p53 protein complexes with circular and linear plasmid DNA substrates suggest involvement of a sliding mechanism.Int J Mol Sci. 2015 Jan 30;16(2):3163-77. doi: 10.3390/ijms16023163. Int J Mol Sci. 2015. PMID: 25647416 Free PMC article.
-
Association of ESR1 germline variants with TP53 somatic variants in breast tumors in a genome-wide study.medRxiv [Preprint]. 2023 Dec 6:2023.12.06.23299442. doi: 10.1101/2023.12.06.23299442. medRxiv. 2023. Update in: Cancer Res Commun. 2024 Jun 27;4(6):1597-1608. doi: 10.1158/2767-9764.CRC-24-0026. PMID: 38106140 Free PMC article. Updated. Preprint.
-
Unaffected Li-Fraumeni Syndrome Carrier Parent Demonstrates Allele-Specific mRNA Stabilization of Wild-Type TP53 Compared to Affected Offspring.Genes (Basel). 2022 Dec 7;13(12):2302. doi: 10.3390/genes13122302. Genes (Basel). 2022. PMID: 36553570 Free PMC article.
-
p53 target gene SMAR1 is dysregulated in breast cancer: its role in cancer cell migration and invasion.PLoS One. 2007 Aug 1;2(7):e660. doi: 10.1371/journal.pone.0000660. PLoS One. 2007. PMID: 17668048 Free PMC article.
-
Battle against cancer: an everlasting saga of p53.Int J Mol Sci. 2014 Dec 1;15(12):22109-27. doi: 10.3390/ijms151222109. Int J Mol Sci. 2014. PMID: 25470027 Free PMC article. Review.
References
-
- Vousden K.H., Lu X. Live or let die: the cell's response to p53. Nature Rev. Cancer. 2002;2:594–604. - PubMed
-
- Sigal A., Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60:6788–6793. - PubMed
-
- Sampath J., Sun D., Kidd V.J., Grenet J., Gandhi A., Shapiro L.H., Wang Q., Zambetti G.P., Schuetz J.D. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J. Biol. Chem. 2001;276:39359–39367. - PubMed
-
- Kim E., Gunther W., Yoshizato K., Meissner H., Zapf S., Nusing R.M., Yamamoto H., Van Meir E.G., Deppert W., Giese A. Tumor suppressor p53 inhibits transcriptional activation of invasion gene thromboxane synthase mediated by the proto-oncogenic factor ets-1. Oncogene. 2003;22:7716–7727. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

