The SNARE Ykt6 is released from yeast vacuoles during an early stage of fusion
- PMID: 15723044
- PMCID: PMC1299260
- DOI: 10.1038/sj.embor.7400350
The SNARE Ykt6 is released from yeast vacuoles during an early stage of fusion
Abstract
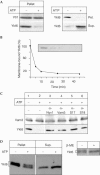
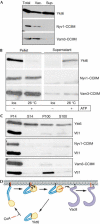
The farnesylated SNARE (N-ethylmaleimide-sensitive factor attachment protein receptor) Ykt6 mediates protein palmitoylation at the yeast vacuole by means of its amino-terminal longin domain. Ykt6 is localized equally to membranes and the cytosol, although it is unclear how this distribution is mediated. We now show that Ykt6 is released efficiently from vacuoles during an early stage of yeast vacuole fusion. This release is dependent on the disassembly of vacuolar SNAREs (priming). In recent literature, it had been demonstrated for mammalian Ykt6 that the membrane-bound form is both palmitoylated and farnesylated at its carboxy-terminal CAAX box, whereas soluble Ykt6 is only farnesylated. In agreement with this, we find that yeast Ykt6 becomes palmitoylated in vitro at its C-terminal CAAX motif. Mutagenesis of the potential palmitoylation site in yeast Ykt6 prevents stable membrane association and is lethal. On the basis of these and other findings, we speculate that Ykt6 is released from membranes by depalmitoylation. Such a mechanism could enable recycling of this lipid-anchored SNARE from the vacuole independent of retrograde transport.
Figures



Similar articles
-
Intramolecular protein-protein and protein-lipid interactions control the conformation and subcellular targeting of neuronal Ykt6.J Cell Sci. 2004 Sep 1;117(Pt 19):4495-508. doi: 10.1242/jcs.01314. J Cell Sci. 2004. PMID: 15331663
-
Ykt6 functionally overlaps with vacuolar and exocytic R-SNAREs in the yeast Saccharomyces cerevisiae.J Biol Chem. 2024 May;300(5):107274. doi: 10.1016/j.jbc.2024.107274. Epub 2024 Apr 6. J Biol Chem. 2024. PMID: 38588809 Free PMC article.
-
ATP-independent control of Vac8 palmitoylation by a SNARE subcomplex on yeast vacuoles.J Biol Chem. 2005 Apr 15;280(15):15348-55. doi: 10.1074/jbc.M410582200. Epub 2005 Feb 8. J Biol Chem. 2005. PMID: 15701652
-
Probing protein palmitoylation at the yeast vacuole.Methods. 2006 Oct;40(2):171-6. doi: 10.1016/j.ymeth.2006.06.020. Methods. 2006. PMID: 17012029 Review.
-
Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles.Annu Rev Cell Dev Biol. 2010;26:115-36. doi: 10.1146/annurev-cellbio-100109-104131. Annu Rev Cell Dev Biol. 2010. PMID: 20521906 Review.
Cited by
-
Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p.EMBO J. 2006 Apr 19;25(8):1579-89. doi: 10.1038/sj.emboj.7601051. Epub 2006 Apr 6. EMBO J. 2006. PMID: 16601699 Free PMC article.
-
Unique self-palmitoylation activity of the transport protein particle component Bet3: a mechanism required for protein stability.Proc Natl Acad Sci U S A. 2006 Aug 22;103(34):12701-6. doi: 10.1073/pnas.0603513103. Epub 2006 Aug 14. Proc Natl Acad Sci U S A. 2006. PMID: 16908848 Free PMC article.
-
Evidence for prenylation-dependent targeting of a Ykt6 SNARE in Plasmodium falciparum.Mol Biochem Parasitol. 2011 Feb;175(2):162-8. doi: 10.1016/j.molbiopara.2010.11.007. Epub 2010 Nov 12. Mol Biochem Parasitol. 2011. PMID: 21075148 Free PMC article.
-
A lipid-anchored SNARE supports membrane fusion.Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17325-30. doi: 10.1073/pnas.1113888108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987819 Free PMC article.
-
Cell biology of protein-lipid conjugation.Cell Struct Funct. 2023 May 11;48(1):99-112. doi: 10.1247/csf.23016. Epub 2023 Apr 6. Cell Struct Funct. 2023. PMID: 37019684 Free PMC article. Review.
References
-
- Bonifacino JS, Glick BS (2004) The mechanisms of vesicle budding and fusion. Cell 116: 153–166 - PubMed
-
- Dietrich LE, Boeddinghaus C, LaGrassa TJ, Ungermann C (2003) Control of eukaryotic membrane fusion by N-terminal domains of SNARE proteins. Biochim Biophys Acta 1641: 111–119 - PubMed
-
- Duncan JA, Gilman AG (2002) Characterization of Saccharomyces cerevisiae acyl-protein thioesterase 1, the enzyme responsible for G protein α subunit deacylation in vivo. J Biol Chem 277: 31740–31752 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

