Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation
- PMID: 15723077
- PMCID: PMC4692364
- DOI: 10.1038/nsmb906
Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation
Abstract
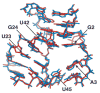
The majority of structural efforts addressing RNA's catalytic function have focused on natural ribozymes, which catalyze phosphodiester transfer reactions. By contrast, little is known about how RNA catalyzes other types of chemical reactions. We report here the crystal structures of a ribozyme that catalyzes enantioselective carbon-carbon bond formation by the Diels-Alder reaction in the unbound state and in complex with a reaction product. The RNA adopts a lambda-shaped nested pseudoknot architecture whose preformed hydrophobic pocket is precisely complementary in shape to the reaction product. RNA folding and product binding are dictated by extensive stacking and hydrogen bonding, whereas stereoselection is governed by the shape of the catalytic pocket. Catalysis is apparently achieved by a combination of proximity, complementarity and electronic effects. We observe structural parallels in the independently evolved catalytic pocket architectures for ribozyme- and antibody-catalyzed Diels-Alder carbon-carbon bond-forming reactions.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures
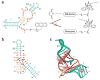
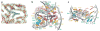
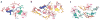

Comment in
-
How RNA closes a Diel.Nat Struct Mol Biol. 2005 Mar;12(3):206-8. doi: 10.1038/nsmb0305-206. Nat Struct Mol Biol. 2005. PMID: 15744318 No abstract available.
Similar articles
-
The structural basis of ribozyme-catalyzed RNA assembly.Science. 2007 Mar 16;315(5818):1549-53. doi: 10.1126/science.1136231. Science. 2007. PMID: 17363667
-
Architecture of a Diels-Alderase ribozyme with a preformed catalytic pocket.Chem Biol. 2004 Sep;11(9):1217-27. doi: 10.1016/j.chembiol.2004.06.011. Chem Biol. 2004. PMID: 15380182
-
Three critical hydrogen bonds determine the catalytic activity of the Diels-Alderase ribozyme.Nucleic Acids Res. 2012 Feb;40(3):1318-30. doi: 10.1093/nar/gkr812. Epub 2011 Oct 5. Nucleic Acids Res. 2012. PMID: 21976731 Free PMC article.
-
Metal ion binding and function in natural and artificial small RNA enzymes from a structural perspective.Met Ions Life Sci. 2011;9:299-345. Met Ions Life Sci. 2011. PMID: 22010277 Review.
-
Ribozyme-catalysed carbon-carbon bond formation.Biochem Soc Trans. 2002 Nov;30(Pt 6):1137-40. doi: 10.1042/bst0301137. Biochem Soc Trans. 2002. PMID: 12440990 Review.
Cited by
-
Structure-based DNA-targeting strategies with small molecule ligands for drug discovery.Med Res Rev. 2013 Sep;33(5):1119-73. doi: 10.1002/med.21278. Epub 2013 Apr 30. Med Res Rev. 2013. PMID: 23633219 Free PMC article. Review.
-
Aptamers, Riboswitches, and Ribozymes in S. cerevisiae Synthetic Biology.Life (Basel). 2021 Mar 17;11(3):248. doi: 10.3390/life11030248. Life (Basel). 2021. PMID: 33802772 Free PMC article. Review.
-
Synthesis of a 2'-Se-thymidine phosphoramidite and its incorporation into oligonucleotides for crystal structure study.Org Lett. 2007 Mar 1;9(5):749-52. doi: 10.1021/ol062937w. Epub 2007 Jan 31. Org Lett. 2007. PMID: 17263541 Free PMC article.
-
The UA_handle: a versatile submotif in stable RNA architectures.Nucleic Acids Res. 2009 Jan;37(1):215-30. doi: 10.1093/nar/gkn911. Epub 2008 Nov 26. Nucleic Acids Res. 2009. PMID: 19036788 Free PMC article.
-
DNA as a versatile chemical component for catalysis, encoding, and stereocontrol.Angew Chem Int Ed Engl. 2010 Sep 24;49(40):7180-201. doi: 10.1002/anie.200906345. Angew Chem Int Ed Engl. 2010. PMID: 20669202 Free PMC article. Review.
References
-
- Kruger K, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence in Tetrahymena. Cell. 1982;31:145–157. - PubMed
-
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. - PubMed
-
- Gilbert W. The RNA world. Nature. 1986;319:618–620.
-
- Cech TR. Ribozymes, the first 20 years. Biochem Soc Trans. 2002;30:1162–1166. - PubMed
-
- Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

