Screening of DNA copy-number aberrations in gastric cancer cell lines by array-based comparative genomic hybridization
- PMID: 15723654
- PMCID: PMC11160020
- DOI: 10.1111/j.1349-7006.2005.00016.x
Screening of DNA copy-number aberrations in gastric cancer cell lines by array-based comparative genomic hybridization
Abstract
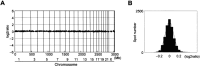
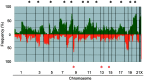
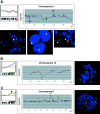
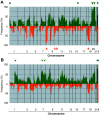
We performed genome-wide screening for deoxyribonucleic acid copy-number aberrations in 31 gastric cancer (GC) cell lines by using custom-made comparative genomic hybridization (CGH)-array. Copy-number gains were frequently detected at 1q, 3q, 5p, 7p, 7q, 8q, 11q, 17q, 20p, 20q, Xp and Xq, and losses at 3p, 4p, 4q, 8p, 9p, 18p and 18q. With respect to histological subtypes, copy-number gains at 1p, 16p, 20p, 20q and 22q, and losses at 8p, 10p, 10q and 18q were significantly frequent in cell lines derived from tumors of the well-differentiated type, whereas copy-number gains at 1q, 7p, 7q, Xp and Xq were frequent in the undifferentiated type. Homozygous deletions were seen at five loci, whereas high-level amplifications were detected in 15 of the 31 GC cell lines; these had occurred at 24 loci, including the segment containing CDK6 (7q21.2). Amplification of that gene had never been reported in GC before. Immunohistochemical studies showed increased levels of CDK6 protein in 54 of the 292 primary GC samples we examined (18.5%). Cytoplasmic localization of CDK6, as well as CDK6 over-expression, was more frequent in well-differentiated GC than in undifferentiated tumors. Nuclear expression of CDK6 was more frequent in early stage GC than in advanced tumors, suggesting that nuclear localization of CDK6 is likely to be a prognostic factor for GC. Taken together, our data indicate that CDK6 might be involved in the pathogenesis of GC and, more generally, that CGH-arrays have a powerful potential for identifying novel cancer-related genetic changes in a variety of tumors.
Figures


Similar articles
-
Genomic aberrations in carcinomas of the uterine corpus.Genes Chromosomes Cancer. 2004 Jul;40(3):229-46. doi: 10.1002/gcc.20038. Genes Chromosomes Cancer. 2004. PMID: 15139002
-
Aberrations of chromosomes 5 and 8 as recurrent cytogenetic events in anaplastic carcinoma of the thyroid as detected by fluorescence in situ hybridisation and comparative genomic hybridisation.Virchows Arch. 2000 Apr;436(4):312-8. doi: 10.1007/s004280050452. Virchows Arch. 2000. PMID: 10834532
-
Distinct chromosomal aberrations of ampulla of Vater and pancreatic head cancers detected by laser capture microdissection and comparative genomic hybridization.Oncol Rep. 2005 Oct;14(4):867-72. Oncol Rep. 2005. PMID: 16142344
-
Cytogenetic aberrations in primary and recurrent fibrolamellar hepatocellular carcinoma detected by comparative genomic hybridization.Am J Clin Pathol. 2000 Dec;114(6):867-74. doi: 10.1309/BMTT-JBPD-D13H-1UVD. Am J Clin Pathol. 2000. PMID: 11338475
-
Genome-wide screening of laser capture microdissected gastric signet-ring cell carcinomas.J Nippon Med Sch. 2002 Jun;69(3):235-42. doi: 10.1272/jnms.69.235. J Nippon Med Sch. 2002. PMID: 12068314 Review.
Cited by
-
MiR-29c is downregulated in gastric carcinomas and regulates cell proliferation by targeting RCC2.Mol Cancer. 2013 Feb 25;12:15. doi: 10.1186/1476-4598-12-15. Mol Cancer. 2013. PMID: 23442884 Free PMC article.
-
Identification of genes with a correlation between copy number and expression in gastric cancer.BMC Med Genomics. 2012 May 4;5:14. doi: 10.1186/1755-8794-5-14. BMC Med Genomics. 2012. PMID: 22559327 Free PMC article.
-
Roles of eukaryotic initiation factor 5A2 in human cancer.Int J Biol Sci. 2013 Oct 12;9(10):1013-20. doi: 10.7150/ijbs.7191. eCollection 2013. Int J Biol Sci. 2013. PMID: 24250246 Free PMC article. Review.
-
Breast cancer cell lines carry cell line-specific genomic alterations that are distinct from aberrations in breast cancer tissues: comparison of the CGH profiles between cancer cell lines and primary cancer tissues.BMC Cancer. 2010 Jan 14;10:15. doi: 10.1186/1471-2407-10-15. BMC Cancer. 2010. PMID: 20070913 Free PMC article.
-
Differential roles of STAT3 depending on the mechanism of STAT3 activation in gastric cancer cells.Br J Cancer. 2011 Jul 26;105(3):407-12. doi: 10.1038/bjc.2011.246. Epub 2011 Jul 5. Br J Cancer. 2011. PMID: 21730976 Free PMC article.
References
-
- Whelan SL, Parkin DM, Masuyer E. Trends in cancer incidence and mortality. Lyon: IARC Scientific, 1993.
-
- Tahara E. Molecular biology of gastric cancer. World J Surg 1995; 19: 484–90. - PubMed
-
- Sakakura C, Mori T, Sakabe T, Ariyama Y, Shinomiya T, Date K, Hagiwara A, Yamaguchi T, Takahashi T, Nakamura Y, Abe T, Inazawa J. Gains, losses, and amplifications of genomic materials in primary gastric cancers analyzed by comparative genomic hybridization. Genes Chromosomes Cancer 1999; 24: 299–305. - PubMed
-
- Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, El‐Rifai W, Hemmer S, Wasenius VM, Vidgren V, Zhu Y. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 1998; 152: 1107–23. - PMC - PubMed
-
- Tay ST, Leong SH, Yu K, Aggarwal A, Tan SY, Lee CH, Wong K, Visvanathan J, Lim D, Wong WK, Soo KC, Kon OL, Tan P. A combined comparative genomic hybridization and expression microarray analysis of gastric cancer reveals novel molecular subtypes. Cancer Res 2003; 63: 3309–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

