Gene duplication, gene loss and evolution of expression domains in the vertebrate nuclear receptor NR5A (Ftz-F1) family
- PMID: 15725073
- PMCID: PMC1184535
- DOI: 10.1042/BJ20050005
Gene duplication, gene loss and evolution of expression domains in the vertebrate nuclear receptor NR5A (Ftz-F1) family
Abstract
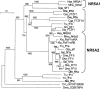
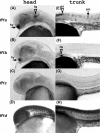
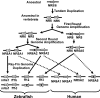
Fushi tarazu factor 1 (Ftz-F1, NR5A) is a zinc-finger transcription factor that belongs to the nuclear receptor superfamily and regulates genes that are involved in sterol and steroid metabolism in gonads, adrenals, liver and other tissues. To understand the evolutionary origins and developmental genetic relationships of the Ftz-F1 genes, we have cloned four homologous Ftz-f1 genes in zebrafish, called ff1a, ff1b, ff1c and ff1d. These four genes have different temporal and spatial expression patterns during development, indicating that they have distinct mechanisms of genetic regulation. Among them, the ff1a expression pattern is similar to mammalian Nr5a2, while the ff1b pattern is similar to that of mammalian Nr5a1. Genetic mapping experiments show that these four ff1 genes are located on chromosome segments conserved between the zebrafish and human genomes, indicating a common ancestral origin. Phylogenetic and conserved synteny analysis show that ff1a is the orthologue of NR5A2, and that ff1b and ff1d genes are co-orthologues of NR5A1 that arose by a gene-duplication event, probably a whole-genome duplication, in the ray-fin lineage, and each gene is located next to an NR6A1 co-orthologue as in humans, showing that the tandem duplication occurred before the divergence of human and zebrafish lineages. ff1c does not have a mammalian counterpart. Thus we have characterized the phylogenetic relationships, expression patterns and chromosomal locations of these Ftz-F1 genes, and have demonstrated their identities as NR5A genes in relation to the orthologous genes in other species.
Figures
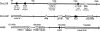


Similar articles
-
Determination of the expression pattern of the dual promoter of zebrafish fushi tarazu factor-1a following microinjections into zebrafish one cell stage embryos.Gen Comp Endocrinol. 2005 May 15;142(1-2):222-6. doi: 10.1016/j.ygcen.2004.12.020. Epub 2005 Feb 5. Gen Comp Endocrinol. 2005. PMID: 15862566
-
Novel steroidogenic factor-1 homolog (ff1d) is coexpressed with anti-Mullerian hormone (AMH) in zebrafish.Dev Dyn. 2005 Jun;233(2):595-604. doi: 10.1002/dvdy.20335. Dev Dyn. 2005. PMID: 15768398
-
Teleost FTZ-F1 homolog and its splicing variant determine the expression of the salmon gonadotropin IIbeta subunit gene.Mol Endocrinol. 1997 Jun;11(7):877-90. doi: 10.1210/mend.11.7.9947. Mol Endocrinol. 1997. PMID: 9178748
-
Zebrafish sex determination and differentiation: involvement of FTZ-F1 genes.Reprod Biol Endocrinol. 2005 Nov 10;3:63. doi: 10.1186/1477-7827-3-63. Reprod Biol Endocrinol. 2005. PMID: 16281973 Free PMC article. Review.
-
Expression of Fushi tarazu factor 1 homolog and Pit-1 genes in the pituitaries of pre-spawning chum and sockeye salmon.Comp Biochem Physiol B Biochem Mol Biol. 2001 Jun;129(2-3):503-9. doi: 10.1016/s1096-4959(01)00348-7. Comp Biochem Physiol B Biochem Mol Biol. 2001. PMID: 11399485 Review.
Cited by
-
Paralogous vitamin D receptors in teleosts: transition of nuclear receptor function.Endocrinology. 2008 May;149(5):2411-22. doi: 10.1210/en.2007-1256. Epub 2008 Feb 7. Endocrinology. 2008. PMID: 18258682 Free PMC article.
-
Disrupted-in-Schizophrenia-1 is essential for normal hypothalamic-pituitary-interrenal (HPI) axis function.Hum Mol Genet. 2017 Jun 1;26(11):1992-2005. doi: 10.1093/hmg/ddx076. Hum Mol Genet. 2017. PMID: 28334933 Free PMC article.
-
Molecular cloning and expression analysis of fushi tarazu factor 1 in the brain of air-breathing catfish, Clarias gariepinus.PLoS One. 2011;6(12):e28867. doi: 10.1371/journal.pone.0028867. Epub 2011 Dec 28. PLoS One. 2011. PMID: 22216130 Free PMC article.
-
DiRE: identifying distant regulatory elements of co-expressed genes.Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W133-9. doi: 10.1093/nar/gkn300. Epub 2008 May 17. Nucleic Acids Res. 2008. PMID: 18487623 Free PMC article.
-
Molecular cloning and sexually dimorphic expression patterns of nr0b1 and nr5a2 in olive flounder, Paralichthys olivaceus.Dev Genes Evol. 2015 Apr;225(2):95-104. doi: 10.1007/s00427-015-0495-2. Epub 2015 Mar 11. Dev Genes Evol. 2015. PMID: 25758177
References
-
- Maglich J. M., Sluder A. E., Willson T. M., Moore J. T. Beyond the human genome: examples of nuclear receptor analysis in model organisms and potential for drug discovery. Am. J. Pharmacogenomics. 2003;3:345–353. - PubMed
-
- Gissendanner C. R., Crossgrove K., Kraus K. A., Maina C. V., Sluder A. E. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev. Biol. 2004;266:399–416. - PubMed
-
- Parker K. L., Rice D. A., Lala D. S., Ikeda Y., Luo X., Wong M., Bakke M., Zhao L., Frigeri C., Hanley N. A., et al. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog. Horm. Res. 2002;57:19–36. - PubMed
-
- Fayard E., Auwerx J., Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. - PubMed
-
- Wolfe K. H. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2001;2:333–341. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

