The tetraspanin D6.1A and its molecular partners on rat carcinoma cells
- PMID: 15725074
- PMCID: PMC1184542
- DOI: 10.1042/BJ20041287
The tetraspanin D6.1A and its molecular partners on rat carcinoma cells
Abstract
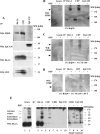
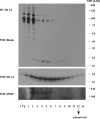
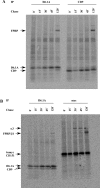
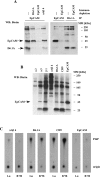
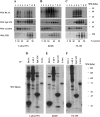
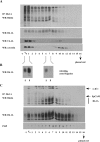
Tetraspanins function as molecular organizers of multi-protein complexes by assembling primary complexes of a relatively low mass into extensive networks involved in cellular signalling. In this paper, we summarize our studies performed on the tetraspanin D6.1A/CO-029/TM4SF3 expressed by rat carcinoma cells. Primary complexes of D6.1A are almost indistinguishable from complexes isolated with anti-CD9 antibody. Indeed, both tetraspanins directly associate with each other and with a third tetraspanin, CD81. Moreover, FPRP (prostaglandin F2alpha receptor-regulatory protein)/EWI-F/CD9P-1), an Ig superfamily member that has been described to interact with CD9 and CD81, is also a prominent element in D6.1A complexes. Primary complexes isolated with D6.1A-specific antibody are clearly different from complexes containing the tetraspanin CD151. CD151 is found to interact only with D6.1A if milder conditions, i.e. lysis with LubrolWX instead of Brij96, are applied to disrupt cellular membranes. CD151 probably mediates the interaction of D6.1A primary complexes with alpha3beta1 integrin. In addition, two other molecules were identified to be part of D6.1A complexes at this higher level of association: type II phosphatidylinositol 4-kinase and EpCAM, an epithelial marker protein overexpressed by many carcinomas. The characterization of the D6.1A core complex and additional more indirect interactions will help to elucidate the role in tumour progression and metastasis attributed to D6.1A.
Figures
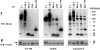

References
-
- Huerta S., Harris D. M., Jazirehi A., Bonavida B., Elashoff D., Livingston E. H., Heber D. Gene expression profile of metastatic colon cancer cells resistant to cisplatin-induced apoptosis. Int. J. Oncol. 2003;22:663–670. - PubMed
-
- Tanaka F., Hori N., Sato K. Identification of differentially expressed genes in rat hepatoma cell lines using subtraction and microarray. J. Biochem. (Tokyo) 2002;131:39–44. - PubMed
-
- Kanetaka K., Sakamoto M., Yamamoto Y., Yamasaki S., Lanza F., Kanematsu T., Hirohashi S. Overexpression of tetraspanin CO-029 in hepatocellular carcinoma. J. Hepatol. 2001;35:637–642. - PubMed
-
- Kanetaka K., Sakamoto M., Yamamoto Y., Takamura M., Kanematsu T., Hirohashi S. Possible involvement of tetraspanin CO-029 in hematogenous intrahepatic metastasis of liver cancer cells. J. Gastroenterol. Hepatol. 2003;18:1309–1314. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

