Mining genetic epidemiology data with Bayesian networks application to APOE gene variation and plasma lipid levels
- PMID: 15725730
- PMCID: PMC1201451
- DOI: 10.1089/cmb.2005.12.1
Mining genetic epidemiology data with Bayesian networks application to APOE gene variation and plasma lipid levels
Abstract
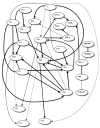
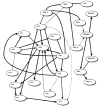
There is a critical need for data-mining methods that can identify SNPs that predict among individual variation in a phenotype of interest and reverse-engineer the biological network of relationships between SNPs, phenotypes, and other factors. This problem is both challenging and important in light of the large number of SNPs in many genes of interest and across the human genome. A potentially fruitful form of exploratory data analysis is the Bayesian or Belief network. A Bayesian or Belief network provides an analytic approach for identifying robust predictors of among-individual variation in a disease endpoints or risk factor levels. We have applied Belief networks to SNP variation in the human APOE gene and plasma apolipoprotein E levels from two samples: 702 African-Americans from Jackson, MS, and 854 non-Hispanic whites from Rochester, MN. Twenty variable sites in the APOE gene were genotyped in both samples. In Jackson, MS, SNPs 4036 and 4075 were identified to influence plasma apoE levels. In Rochester, MN, SNPs 3937 and 4075 were identified to influence plasma apoE levels. All three SNPs had been previously implicated in affecting measures of lipid and lipoprotein metabolism. Like all data-mining methods, Belief networks are meant to complement traditional hypothesis-driven methods of data analysis. These results document the utility of a Belief network approach for mining large scale genotype-phenotype association data.
Figures
References
-
- Friedman, N., Goldszmidt, M., and Wyner, A. 1999. Data analysis with Bayesian networks: A bootstrap approach. Proc. 15th Conf. on Uncertainty in Artificial Intelligence, UAI, 196–205.
-
- Friedman N, Linial M, Nachman I, Pe’er D. Using Bayesian networks to analyze expression data. J Comp Biol. 2000;7:601–620. - PubMed
-
- Han, J., and Kamber, M. 2001. Data Mining: Concepts and Techniques, Morgan Kaufmann, San Francisco, CA.
-
- Heckerman, D. 1995. A tutorial on learning with Bayesian networks. Technical report MSR-TR-95-06, Microsoft Research.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

