Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications
- PMID: 15728358
- PMCID: PMC549290
- DOI: 10.1073/pnas.0409851102
Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications
Abstract
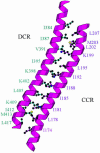
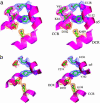
Type I restriction-modification enzymes are differentiated from type II and type III enzymes by their recognition of two specific dsDNA sequences separated by a given spacer and cleaving DNA randomly away from the recognition sites. They are oligomeric proteins formed by three subunits: a specificity subunit, a methylation subunit, and a restriction subunit. We solved the crystal structure of a specificity subunit from Methanococcus jannaschii at 2.4-A resolution. Two highly conserved regions (CRs) in the middle and at the C terminus form a coiled-coil of long antiparallel alpha-helices. Two target recognition domains form globular structures with almost identical topologies and two separate DNA binding clefts with a modeled DNA helix axis positioned across the CR helices. The structure suggests that the coiled-coil CRs act as a molecular ruler for the separation between two recognized DNA sequences. Furthermore, the relative orientation of the two DNA binding clefts suggests kinking of bound dsDNA and exposing of target adenines from the recognized DNA sequences.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

