Quantitative kinetic analysis of the bacteriophage lambda genetic network
- PMID: 15728384
- PMCID: PMC549295
- DOI: 10.1073/pnas.0500670102
Quantitative kinetic analysis of the bacteriophage lambda genetic network
Abstract
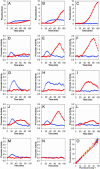
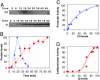
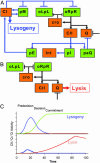
The lysis-lysogeny decision of bacteriophage lambda has been a paradigm for a developmental genetic network, which is composed of interlocked positive and negative feedback loops. This genetic network is capable of responding to environmental signals and to the number of infecting phages. An interplay between CI and Cro functions suggested a bistable switch model for the lysis-lysogeny decision. Here, we present a real-time picture of the execution of lytic and lysogenic pathways with unprecedented temporal resolution. We monitor, in vivo, both the level and function of the CII and Q gene regulators. These activators are cotranscribed yet control opposite developmental pathways. Conditions that favor the lysogenic response show severe delay and down-regulation of Q activity, in both CII-dependent and CII-independent ways. Whereas CII activity correlates with its protein level, Q shows a pronounced threshold before its function is observed. Our quantitative analyses suggest that by regulating CII and CIII, Cro plays a key role in the ability of the lambda genetic network to sense the difference between one and more than one phage particles infecting a cell. Thus, our results provide an improved framework to explain the longstanding puzzle of the decision process.
Figures

Comment in
-
Threshold effects in gene regulation: when some is not enough.Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5310-1. doi: 10.1073/pnas.0501645102. Epub 2005 Apr 5. Proc Natl Acad Sci U S A. 2005. PMID: 15811940 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

