Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast
- PMID: 15728720
- PMCID: PMC1087238
- DOI: 10.1091/mbc.e04-11-0996
Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast
Abstract
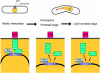
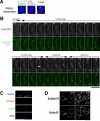
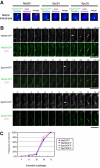
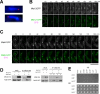
In the fission yeast Schizosaccharomyces pombe, centromeres remain clustered at the spindle-pole body (SPB) during mitotic interphase. In contrast, during meiotic prophase centromeres dissociate from the SPB. Here we examined the behavior of centromere proteins in living meiotic cells of S. pombe. We show that the Nuf2-Ndc80 complex proteins (Nuf2, Ndc80, Spc24, and Spc25) disappear from the centromere in meiotic prophase when the centromeres are separated from the SPB. The centromere protein Mis12 also dissociates during meiotic prophase; however, Mis6 remains throughout meiosis. When cells are induced to meiosis by inactivation of Pat1 kinase (a key negative regulator of meiosis), centromeres remain associated with the SPB during meiotic prophase. However, inactivation of Nuf2 by a mutation causes the release of centromeres from the SPB in pat1 mutant cells, suggesting that the Nuf2-Ndc80 complex connects centromeres to the SPB. We further found that removal of the Nuf2-Ndc80 complex from the centromere and centromere-SPB dissociation are caused by mating pheromone signaling. Because pat1 mutant cells also show aberrant chromosome segregation in the first meiotic division and this aberration is compensated by mating pheromone signaling, dissociation of the Nuf2-Ndc80 complex may be associated with remodeling of the kinetochore for meiotic chromosome segregation.
Figures



References
-
- Appelgren, H., Kniola, B., and Ekwall, K. (2003). Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 116, 4035–4042. - PubMed
-
- Beach, D., Rodgers, L., and Gould, J. (1985). RAN1+ controls the transition from mitotic division to meiosis in fission yeast. Curr. Genet. 10, 297–311. - PubMed
-
- Bernard, P., Maure, J. F., and Javerzat, J. P. (2001). Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat. Cell Biol. 3, 522–526. - PubMed
-
- Bharadwaj, R., Qi, W., and Yu, H. (2004). Identification of two novel components of the human NDC80 kinetochore complex. J. Biol. Chem. 279, 13076–13085. - PubMed
-
- Chikashige, Y., Ding, D. Q., Funabiki, H., Haraguchi, T., Mashiko, S., Yanagida, M., and Hiraoka, Y. (1994). Telomere-led premeiotic chromosome movement in fission yeast. Science 264, 270–273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

